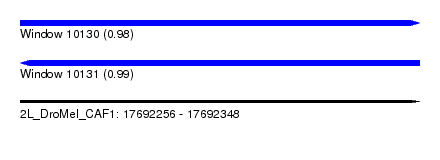
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,692,256 – 17,692,348 |
| Length | 92 |
| Max. P | 0.986394 |

| Location | 17,692,256 – 17,692,348 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17692256 92 + 22407834 CCUUCUUCUGCCUUGUUGCAUGCCACAUGAAAAAUUUGCGUUGACAACAAUGCGACAACAUGUCGUGGCUGGUCCCCCA------GCCCAAAAAGGUA ........((((((.(((...((.(((((......((((((((....))))))))...))))).))((((((....)))------)))))).)))))) ( -29.30) >DroGri_CAF1 92055 78 + 1 GUUGCCUUUGCCUUGUUGCAUGUGGCACGGA-AAUUUGCGUUGACAACAAUGCGAA---AUGUUCUC--UGUUACCGC--------------AAAGUA .((((...(((......))).((((((.(((-(.(((((((((....)))))))))---...)))).--)))))).))--------------)).... ( -22.70) >DroSec_CAF1 83069 92 + 1 CCUUCUUCUGCCUUGUUGCAUGCCACAUGUAAAAUUUGCGUUGACAACAAUGCGACAACAUGUCGUGGCUGGUCUCCCA------GCCAAAAAAGGUA ........((((((.(((((((...))))))).....((((((....))))))(((.....))).(((((((....)))------))))...)))))) ( -30.40) >DroSim_CAF1 65636 92 + 1 CCUUCUUCUGCCUUGUUGCAUGCCACAUGAAAAAUUUGCGUUGACAACAAUGCGACAACAUGUCGUGGCUGGUCUCCCA------GCCAAAAAAGGUA ........((((((((((((((...))))......((((((((....))))))))))))......(((((((....)))------))))...)))))) ( -29.20) >DroYak_CAF1 86849 98 + 1 CCUUCUUCUGCCUUGUUGCAUGCCACAUGAAAAAUUUGCGUUGACAACAAUGCGACAACAUGUCGUGGCUGGUCUCCCGUCUCCCACCCAAAAAGGUA ........((((((.(((...(((((...........((((((....))))))(((.....)))))))).((....))..........))).)))))) ( -26.00) >consensus CCUUCUUCUGCCUUGUUGCAUGCCACAUGAAAAAUUUGCGUUGACAACAAUGCGACAACAUGUCGUGGCUGGUCUCCCA______GCCAAAAAAGGUA ........((((((.......(((((...........((((((....))))))(((.....)))))))).((....))..............)))))) (-18.48 = -18.52 + 0.04)

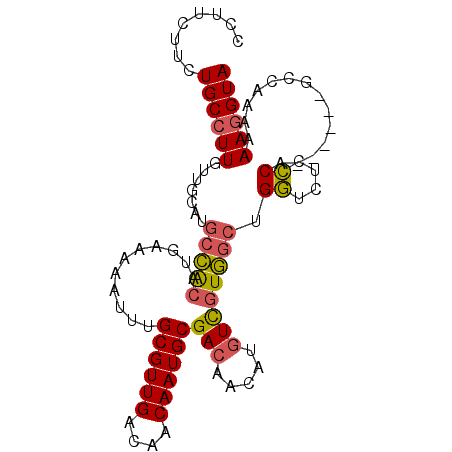
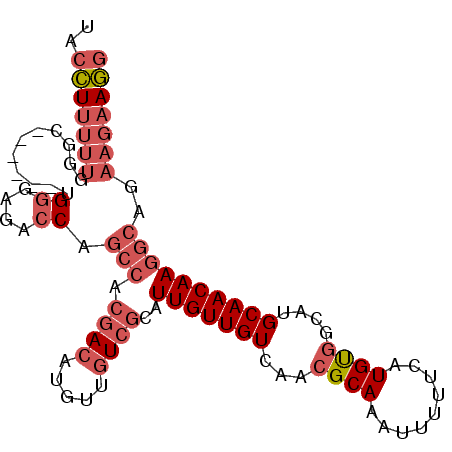
| Location | 17,692,256 – 17,692,348 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -18.36 |
| Energy contribution | -20.48 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17692256 92 - 22407834 UACCUUUUUGGGC------UGGGGGACCAGCCACGACAUGUUGUCGCAUUGUUGUCAACGCAAAUUUUUCAUGUGGCAUGCAACAAGGCAGAAGAAGG ..(((((((.(((------(((....))))))..(((.....)))((.(((((((...((((.........))))....))))))).))..))))))) ( -36.30) >DroGri_CAF1 92055 78 - 1 UACUUU--------------GCGGUAACA--GAGAACAU---UUCGCAUUGUUGUCAACGCAAAUU-UCCGUGCCACAUGCAACAAGGCAAAGGCAAC ...(((--------------(((......--((.((((.---.......)))).))..))))))..-....((((...(((......)))..)))).. ( -18.00) >DroSec_CAF1 83069 92 - 1 UACCUUUUUUGGC------UGGGAGACCAGCCACGACAUGUUGUCGCAUUGUUGUCAACGCAAAUUUUACAUGUGGCAUGCAACAAGGCAGAAGAAGG ..(((((((((((------(((....))))))..(((.....)))((.(((((((...((((.........))))....))))))).)).)))))))) ( -37.50) >DroSim_CAF1 65636 92 - 1 UACCUUUUUUGGC------UGGGAGACCAGCCACGACAUGUUGUCGCAUUGUUGUCAACGCAAAUUUUUCAUGUGGCAUGCAACAAGGCAGAAGAAGG ..(((((((((((------(((....))))))..(((.....)))((.(((((((...((((.........))))....))))))).)).)))))))) ( -37.50) >DroYak_CAF1 86849 98 - 1 UACCUUUUUGGGUGGGAGACGGGAGACCAGCCACGACAUGUUGUCGCAUUGUUGUCAACGCAAAUUUUUCAUGUGGCAUGCAACAAGGCAGAAGAAGG ..(((((((.....(....)((....)).(((.((((.....))))..(((((((...((((.........))))....))))))))))..))))))) ( -30.70) >consensus UACCUUUUUGGGC______UGGGAGACCAGCCACGACAUGUUGUCGCAUUGUUGUCAACGCAAAUUUUUCAUGUGGCAUGCAACAAGGCAGAAGAAGG ..(((((((...........((....)).(((.((((.....))))..(((((((...((((.........))))....))))))))))..))))))) (-18.36 = -20.48 + 2.12)

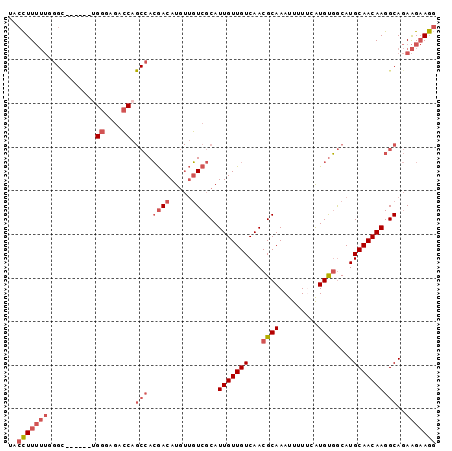
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:40 2006