
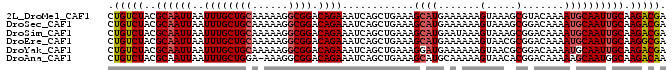
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,678,650 – 17,678,767 |
| Length | 117 |
| Max. P | 0.917444 |

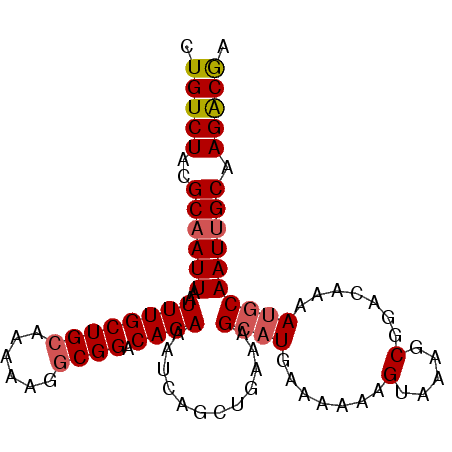
| Location | 17,678,650 – 17,678,743 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17678650 93 - 22407834 CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAAAGCGUACAAAAUGCAAUUGCAAGACGA .(((((..((((((..((((((((......)))).))))..(((((.....)).)))..........((((.....)))))))))).))))). ( -22.40) >DroSec_CAF1 74137 93 - 1 CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAAAGCGGACAAAAUGCAAUUGCAAGACGA .(((((.(((......((((((((......)))).))))..(((((.....)).)))..........))).......(((....)))))))). ( -21.60) >DroSim_CAF1 56678 93 - 1 CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAUAAAGUAAAGCGGACAAAAUGCAAUUGCAAGACGA .(((((.(((......((((((((......)))).))))..(((((.....)).)))..........))).......(((....)))))))). ( -21.60) >DroEre_CAF1 61374 93 - 1 CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAACGCGGACAAAAUGCAAUUGCAAGGCGA .(((((.(((......((((((((......)))).))))..(((((.....)).)))..........))).......(((....)))))))). ( -21.10) >DroYak_CAF1 77784 93 - 1 CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGGAUGAAAAAAGUAACGCGGACAAAAUGCAAUUGCAAGACGA .(((((.(((......((((((((......)))).))))..(((.((....)).)))..........))).......(((....)))))))). ( -22.10) >DroAna_CAF1 55173 92 - 1 CUGUCUACGCAAUUAAUUUGCUGGA-AAAGGCGGACAGAAAUCAGCUGAAAGCAUGCAAAAAGUAACACGGACAAAAAGCAAUGGCAAGACAA .(((((..(((.....((((((...-...)))))).........((.....)).))).....................((....)).))))). ( -15.50) >consensus CUGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAAAGCGGACAAAAUGCAAUUGCAAGACGA .(((((..((((((..((((((((......)))).))))............((((.......(.....).......)))))))))).))))). (-17.47 = -17.86 + 0.39)
| Location | 17,678,669 – 17,678,767 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17678669 98 - 22407834 --------AGUAGUUUCU--------CAGUUUCCUGUCUGC------UGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAAAGCGUACA --------.(((((((..--------(......((((((((------(......((((.....))))........)))))))))...(((((.....)).))).....)..)))).))). ( -24.84) >DroSim_CAF1 56697 98 - 1 --------AGUAGUUUCU--------CAGUUUCCUGUCUGC------UGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAUAAAGUAAAGCGGACA --------....((((.(--------(((((..((((((((------(......((((.....))))........))))))))).....))))))))))..................... ( -24.54) >DroEre_CAF1 61393 98 - 1 --------AGUAGUUUCU--------CAGCUUCCUGUCUGC------UGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAACGCGGACA --------....((((.(--------(((((..((((((((------(......((((.....))))........))))))))).....))))))))))..................... ( -26.94) >DroYak_CAF1 77803 106 - 1 --------AGUAGUUUCUCAGUUUCUCAGUUUCCCGUCUGC------UGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGGAUGAAAAAAGUAACGCGGACA --------...........................((((((------(((((..((((.....)))).((......)))))))....(((.((....)).)))..........)))))). ( -23.30) >DroMoj_CAF1 82427 88 - 1 --------AGCAGUUAC---------CCGUUGCCUGUCUGCUCCGCCUGUCUGCGCAAUUAAUUUGCAGCA--AAGGCGGACACAAAUCAGCUG-------------CGUAACACGAACU --------....(((((---------(.((((......((.(((((((...(((((((.....)))).)))--.))))))))).....)))).)-------------.)))))....... ( -29.40) >DroAna_CAF1 55192 105 - 1 CAGUUCUGGGUAGUUUUU--------CAGCAUCCUGUCGGC------UGUCUACGCAAUUAAUUUGCUGGA-AAAGGCGGACAGAAAUCAGCUGAAAGCAUGCAAAAAGUAACACGGACA ..(((((..(((..((((--------((((...(((((.((------(.((((.((((.....))))))))-...))).)))))......))))))))..)))....)).)))....... ( -33.50) >consensus ________AGUAGUUUCU________CAGUUUCCUGUCUGC______UGUCUACGCAAUUAAUUUGCUGCAAAAAGGCGGACAGAAAUCAGCUGAAAGCAUGAAAAAAGUAACGCGGACA ..........................(((((..((((((((......(((....((((.....)))).))).....)))))))).....))))).......................... (-15.32 = -16.10 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:35 2006