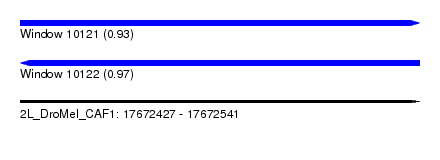
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,672,427 – 17,672,541 |
| Length | 114 |
| Max. P | 0.965505 |

| Location | 17,672,427 – 17,672,541 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -20.49 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
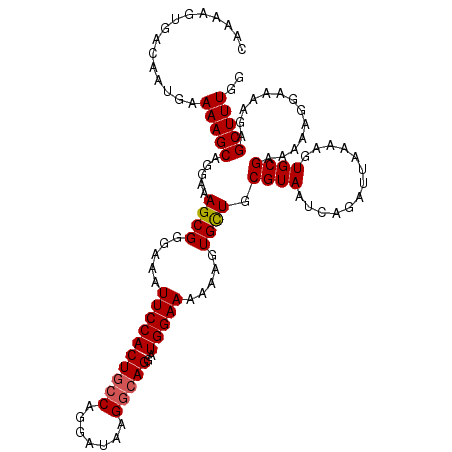
>2L_DroMel_CAF1 17672427 114 + 22407834 CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGGAUAAGACAGGAUGGAAAAAAGUGCUGCGUAAUCAGAUUAAAAGUGCGAAAAAGGAAAAGAGCUUUGG .......................(((((......(((((...((.(.......).)).)))))....(..((...((((...))))..))..)..............))))).. ( -15.40) >DroSec_CAF1 67940 114 + 1 CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGGAUAAGGCAGGAUGGAAAAAAGUGUUGCGUAAUCAGAUUAAAAGUGCGAAAAAGGAAAAGAGCUUUGG .....(..(((.......((......))......((((((((((.......)))))..))))).....)))..)............(((((.(............).))))).. ( -21.20) >DroSim_CAF1 50457 114 + 1 CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGGAUAAGGCAGGAUGGAAAAAAGUGCUGCGUAAUCAGAUUAAAAGUGCGAAAAAGGAAGAGAGCUUUGG .......................(((((......((((.(((((.......)))))...........(..((...((((...))))..))..)......))))....))))).. ( -22.60) >DroEre_CAF1 55298 111 + 1 CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGAAUAAGGCAGCAUGGAA--AACUGCUACGUAAUCAGAUUAAAAGUGCGAAAAAG-AAAAGAGCUUUGG ...............(((((.....(((((....((((((((((.......)))))..)))))--..))))).((((.............))))......-......))))).. ( -24.92) >DroYak_CAF1 71709 112 + 1 CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGGAUAGGGGAGGAUGGAAA-AAUUGCUACGUAAUCAGAUUAAAAGUGCGAAAAAG-AAAAAAGCUUUGG ...............(((((.....(((((....(((((((.((.......)).))..))))).-..))))).((((.............))))......-......))))).. ( -18.32) >consensus CAAAAGUGACAAUGAAAAGCAGGAAAGCGGGAAAUUCCACUGCCAGGAUAAGGCAGGAUGGAAAAAAGUGCUGCGUAAUCAGAUUAAAAGUGCGAAAAAGGAAAAGAGCUUUGG ...............(((((.....((((.....((((((((((.......)))))..))))).....)))).((((.............)))).............))))).. (-18.30 = -18.54 + 0.24)

| Location | 17,672,427 – 17,672,541 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -16.68 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
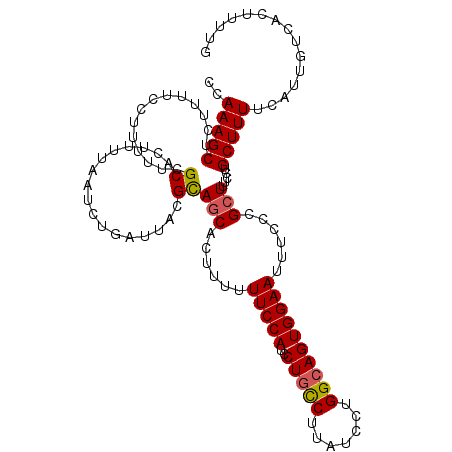
>2L_DroMel_CAF1 17672427 114 - 22407834 CCAAAGCUCUUUUCCUUUUUCGCACUUUUAAUCUGAUUACGCAGCACUUUUUUCCAUCCUGUCUUAUCCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG ..(((((..............((.................))(((......(((((..(((((.......))))))))))......))).....)))))............... ( -16.03) >DroSec_CAF1 67940 114 - 1 CCAAAGCUCUUUUCCUUUUUCGCACUUUUAAUCUGAUUACGCAACACUUUUUUCCAUCCUGCCUUAUCCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG ..(((((..............((.................)).........(((((..(((((.......))))))))))......)))))....................... ( -16.63) >DroSim_CAF1 50457 114 - 1 CCAAAGCUCUCUUCCUUUUUCGCACUUUUAAUCUGAUUACGCAGCACUUUUUUCCAUCCUGCCUUAUCCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG ..(((((..............((.................))(((......(((((..(((((.......))))))))))......))).....)))))............... ( -18.73) >DroEre_CAF1 55298 111 - 1 CCAAAGCUCUUUU-CUUUUUCGCACUUUUAAUCUGAUUACGUAGCAGUU--UUCCAUGCUGCCUUAUUCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG .(((((.......-..........................((((((...--.....)))))).......(((((((((((......((......)).))))))))))).))))) ( -19.40) >DroYak_CAF1 71709 112 - 1 CCAAAGCUUUUUU-CUUUUUCGCACUUUUAAUCUGAUUACGUAGCAAUU-UUUCCAUCCUCCCCUAUCCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG .(((((((....(-(...................))......)))....-...................(((((((((((......((......)).)))))))))))..)))) ( -12.61) >consensus CCAAAGCUCUUUUCCUUUUUCGCACUUUUAAUCUGAUUACGCAGCACUUUUUUCCAUCCUGCCUUAUCCUGGCAGUGGAAUUUCCCGCUUUCCUGCUUUUCAUUGUCACUUUUG ..(((((..............((.................))(((......(((((..(((((.......))))))))))......))).....)))))............... (-14.75 = -14.75 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:32 2006