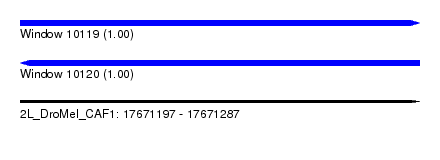
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,671,197 – 17,671,287 |
| Length | 90 |
| Max. P | 0.999923 |

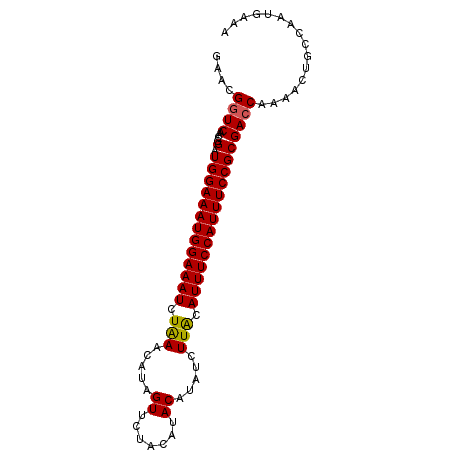
| Location | 17,671,197 – 17,671,287 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17671197 90 + 22407834 UUUCAUUGGCAGUUUUGGUCGCGGAAAUGGAAAUGUAAGAUAUGUAUGUAGAACUAUGUUCAGAUUUCCAUUUCCAUCCGUUGACCGUUC ................(((((((((..((((((((..((((.((.(((((....))))).)).)))).)))))))))))).))))).... ( -25.20) >DroSec_CAF1 66740 90 + 1 UUUCAUUGGCAGUUUUGGUCGCGGAAAUGGAAAUGUAAGAUAUGUAUGUAGAACUGUGUUUAGAUUUCCAUUUCCAUCCGUUGACCGUUC ................((((((((((((((((((.((((....((.......))....)))).)))))))))....)))).))))).... ( -24.70) >DroSim_CAF1 49257 90 + 1 UUUCAUUGGCAGUUUUGUUCGCGGAAAUGGAAAUGUAAGAUAUGUAUGUAGAACUGUGUUUAGAUUUCCAUUUCCAUCCGUUGACCGUUC ..(((.(((..((.......))((((((((((((.((((....((.......))....)))).))))))))))))..))).)))...... ( -22.30) >DroEre_CAF1 54092 90 + 1 UUUCAUUGGCAGUUUUGGUCGCGGAAAUGGAAAUGUAAGAUAUGUAUGUAUAACAAUGUUUAGAUUUCCAUUUCCAUCCGUUGACCGUUC ................((((((((((((((((((.((((...(((.......)))...)))).)))))))))....)))).))))).... ( -25.70) >DroYak_CAF1 70506 90 + 1 UUUCAUAGGCAGUUUUGGUCGCGGAAAUGGAAAUGCAAGAUAUGUAUGUGAAACUAUGUUUAGAUUUCCAUUUCCAUCCGUUGACCGUUC ................((((((((((((((((((.(.((((((((.......)).)))))).))))))))))....)))).))))).... ( -23.90) >consensus UUUCAUUGGCAGUUUUGGUCGCGGAAAUGGAAAUGUAAGAUAUGUAUGUAGAACUAUGUUUAGAUUUCCAUUUCCAUCCGUUGACCGUUC ................((((((((((((((((((.((((....((.......))....)))).)))))))))....)))).))))).... (-21.74 = -22.34 + 0.60)

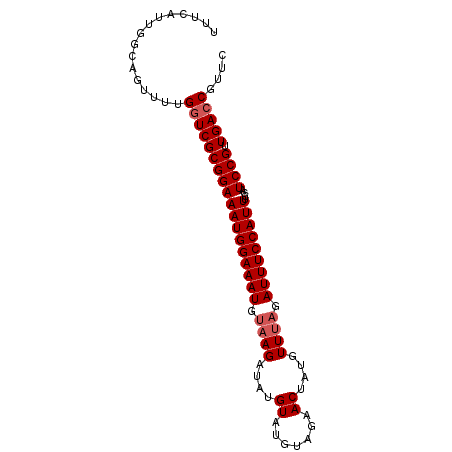
| Location | 17,671,197 – 17,671,287 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17671197 90 - 22407834 GAACGGUCAACGGAUGGAAAUGGAAAUCUGAACAUAGUUCUACAUACAUAUCUUACAUUUCCAUUUCCGCGACCAAAACUGCCAAUGAAA ....((((..(((((((((((((....).((((...))))...............))))))))..)))).))))................ ( -20.60) >DroSec_CAF1 66740 90 - 1 GAACGGUCAACGGAUGGAAAUGGAAAUCUAAACACAGUUCUACAUACAUAUCUUACAUUUCCAUUUCCGCGACCAAAACUGCCAAUGAAA ....((((....(.(((((((((((((.(((.....((.......)).....))).))))))))))))))))))................ ( -20.10) >DroSim_CAF1 49257 90 - 1 GAACGGUCAACGGAUGGAAAUGGAAAUCUAAACACAGUUCUACAUACAUAUCUUACAUUUCCAUUUCCGCGAACAAAACUGCCAAUGAAA ...((.....))...((((((((((((.(((.....((.......)).....))).))))))))))))(((........)))........ ( -17.20) >DroEre_CAF1 54092 90 - 1 GAACGGUCAACGGAUGGAAAUGGAAAUCUAAACAUUGUUAUACAUACAUAUCUUACAUUUCCAUUUCCGCGACCAAAACUGCCAAUGAAA ....((((....(.(((((((((((((.(((....(((.......)))....))).))))))))))))))))))................ ( -22.40) >DroYak_CAF1 70506 90 - 1 GAACGGUCAACGGAUGGAAAUGGAAAUCUAAACAUAGUUUCACAUACAUAUCUUGCAUUUCCAUUUCCGCGACCAAAACUGCCUAUGAAA ....((((..((((((((((((((((.(((....))))))).....((.....))))))))))..)))).))))................ ( -22.50) >consensus GAACGGUCAACGGAUGGAAAUGGAAAUCUAAACAUAGUUCUACAUACAUAUCUUACAUUUCCAUUUCCGCGACCAAAACUGCCAAUGAAA ....((((....(.(((((((((((((.(((.....((.......)).....))).))))))))))))))))))................ (-19.08 = -18.96 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:30 2006