| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,637,530 – 17,637,634 |
| Length | 104 |
| Max. P | 0.930665 |

| Location | 17,637,530 – 17,637,634 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
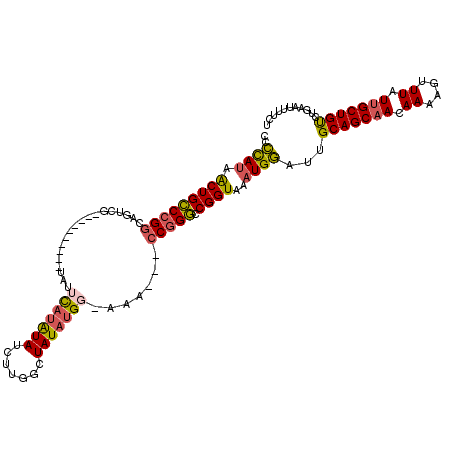
>2L_DroMel_CAF1 17637530 104 + 22407834 AGAAAAUUCAGACAGCAAUAAACUUUUGUUGCUGCAAUCCAUUUACCGACCCGG---UUUGCCAUAUAGCCAAGAUAUAUGAAUA----------CGACUGCCGGGCAGUUAUGGGG ..........(.(((((((((....))))))))))..(((((..((.(.(((((---((((.((((((.......))))))....----------)))..))))))).)).))))). ( -29.90) >DroSec_CAF1 33110 104 + 1 AGAAAUUUCAGACAGCAAUAAACUUUUGUUGCUGCAAUCCAUUUACCGACCCGG---CUUACCAUAUAGCCAAGAUAUAUGAAUA----------CGACUGCCGGGCAGUUAUGGGG ..........(.(((((((((....))))))))))..(((((..((.(.(((((---(....((((((.......))))))....----------.....))))))).)).))))). ( -30.62) >DroEre_CAF1 20724 101 + 1 --AAAAUUCAGACAGCAAUAAACUUUUGUUGCUGCAAUCCAUUUACCGGCCCGG---CUU-GCAUAUAGCCAAGAUAUAUGAAUA----------CGACUGCCGGGCAGUUAUGGGG --........(.(((((((((....))))))))))..(((((..((..((((((---(..-.((((((.......))))))....----------.....))))))).)).))))). ( -33.90) >DroYak_CAF1 34037 103 + 1 AGAAAACUCAGACAGCAAUAAACUUUUGUUGCUGCAAUCCAUUUACCGGCCCGG---UUG-CCAUAUAGCCAAGAUAUAUGAAUA----------CGAAUGCCGGGCAGUUAUGGGG ..........(.(((((((((....))))))))))..(((((..((..((((((---(..-(((((((.......))))))....----------.)...))))))).)).))))). ( -31.70) >DroAna_CAF1 14397 115 + 1 --AAGGAGCAGGCAGCCAUAAACUUUUGUUGCUGCAAUUCAUUUACCGGGCUGCGAAAAUGCCUUAUAGCCUGGAUACGUAUGUGAGAGAGUGGCAGAGUGCCGGCCAGCUGUAAGG --.........((((((.((((...((((....))))....))))...)))))).......(((((((((.(((.(((....)))......((((.....)))).)))))))))))) ( -34.30) >consensus AGAAAAUUCAGACAGCAAUAAACUUUUGUUGCUGCAAUCCAUUUACCGGCCCGG___CUU_CCAUAUAGCCAAGAUAUAUGAAUA__________CGACUGCCGGGCAGUUAUGGGG ..........(.(((((((((....))))))))))..(((((......((((((........((((((.......))))))....................))))))....))))). (-20.69 = -21.17 + 0.48)


| Location | 17,637,530 – 17,637,634 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17637530 104 - 22407834 CCCCAUAACUGCCCGGCAGUCG----------UAUUCAUAUAUCUUGGCUAUAUGGCAAA---CCGGGUCGGUAAAUGGAUUGCAGCAACAAAAGUUUAUUGCUGUCUGAAUUUUCU ..((((.(((((((((..((((----------((((((.......)))..)))))))...---))))).))))..))))...(((((((.((....)).)))))))........... ( -30.30) >DroSec_CAF1 33110 104 - 1 CCCCAUAACUGCCCGGCAGUCG----------UAUUCAUAUAUCUUGGCUAUAUGGUAAG---CCGGGUCGGUAAAUGGAUUGCAGCAACAAAAGUUUAUUGCUGUCUGAAAUUUCU ..((((.((((((((((..(((----------((((((.......)))..))))))...)---))))).))))..))))...(((((((.((....)).)))))))........... ( -30.50) >DroEre_CAF1 20724 101 - 1 CCCCAUAACUGCCCGGCAGUCG----------UAUUCAUAUAUCUUGGCUAUAUGC-AAG---CCGGGCCGGUAAAUGGAUUGCAGCAACAAAAGUUUAUUGCUGUCUGAAUUUU-- ..((((.((((((((((...((----------((((((.......)))..))))).-..)---))))).))))..))))...(((((((.((....)).))))))).........-- ( -30.00) >DroYak_CAF1 34037 103 - 1 CCCCAUAACUGCCCGGCAUUCG----------UAUUCAUAUAUCUUGGCUAUAUGG-CAA---CCGGGCCGGUAAAUGGAUUGCAGCAACAAAAGUUUAUUGCUGUCUGAGUUUUCU ..((((.(((((((((...(((----------((((((.......)))..))))))-...---))))).))))..))))...(((((((.((....)).)))))))........... ( -27.40) >DroAna_CAF1 14397 115 - 1 CCUUACAGCUGGCCGGCACUCUGCCACUCUCUCACAUACGUAUCCAGGCUAUAAGGCAUUUUCGCAGCCCGGUAAAUGAAUUGCAGCAACAAAAGUUUAUGGCUGCCUGCUCCUU-- (((((.((((((..(((.....)))........((....))..))).))).))))).......((((((..((((((...(((......)))..)))))))))))).........-- ( -29.00) >consensus CCCCAUAACUGCCCGGCAGUCG__________UAUUCAUAUAUCUUGGCUAUAUGG_AAA___CCGGGCCGGUAAAUGGAUUGCAGCAACAAAAGUUUAUUGCUGUCUGAAUUUUCU ..((((.(((((((((...................(((((((.......))))))).......))))).))))..))))...(((((((.((....)).)))))))........... (-19.07 = -19.23 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:25 2006