
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
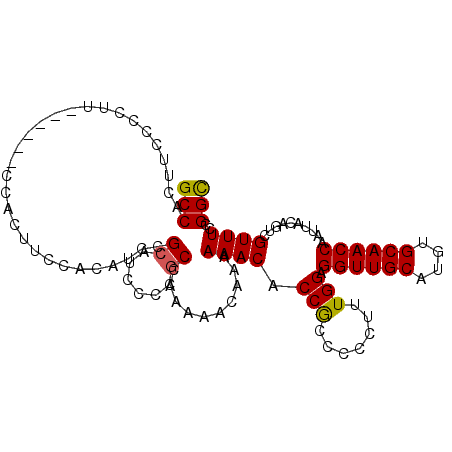
| Location | 17,617,755 – 17,617,895 |
| Length | 140 |
| Max. P | 0.800692 |

| Location | 17,617,755 – 17,617,855 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.07 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -15.92 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
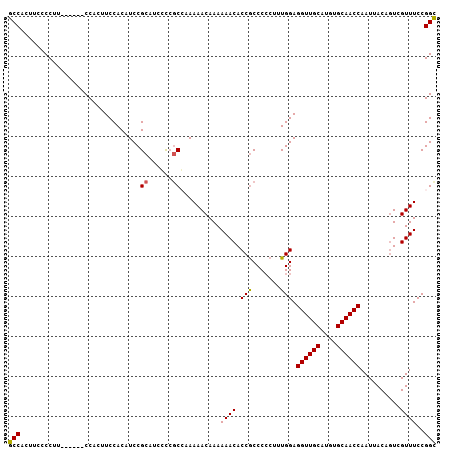
>2L_DroMel_CAF1 17617755 100 + 22407834 GCCACUUCCCCUU------CCACUUCCACAUCCGCAUCAUCGCCAAAAACAAAAAACACCGCCC-CUUUGGAGGUUGCAUGUGCAACCAAUUACAGUCGUUUCCGGC .............------......................(((..((((........(((...-...))).((((((....))))))..........))))..))) ( -17.60) >DroSec_CAF1 13146 101 + 1 GCCACUUCCCCUU------CCACUUCCACAUCCGCAUCGCCGCCAAAAACAAAAAACACCGCCCCCAUUGGAGGUUGCAUGUGCAACCAAUUACAGUCGUUUCCGGU .............------...................((((..((..((........(((.......))).((((((....)))))).......))..))..)))) ( -16.00) >DroSim_CAF1 3335 101 + 1 GCCACUUCCCCUU------CCACUUCCACAUCCGCAUCGCCGCCAAAAACAAAAAACACCGCCCCCAUUGGAGGUUGCAUGUGCAACCAAUUACAUUCGUUUCCGGC .............------...................((((....((((........(((.......))).((((((....))))))..........)))).)))) ( -17.40) >DroEre_CAF1 1565 100 + 1 GCCACUUCCCCUU------CCACUUCCACUUCCGCAUCCCAGCGAAAAACAAAAAACACCGCCCCCU-UGGAGGUUGCAUGUGCAACCAAUUACAGUCGUUUCCGGC (((..........------.............(((......))).........((((.(((......-))).((((((....))))))..........))))..))) ( -19.80) >DroYak_CAF1 13090 107 + 1 GCCACUUCCCCUUCUCAUGCCACUUCCACUUCCGCAUCCCCACGAAAAACAAAAAACACCACCCCCUUUGGAGGUUGCAUGUGCAACCAAUUACAGUCGUUUCCGGC (((.............((((.............))))....((((.............(((.......))).((((((....))))))........))))....))) ( -18.82) >consensus GCCACUUCCCCUU______CCACUUCCACAUCCGCAUCCCCGCCAAAAACAAAAAACACCGCCCCCUUUGGAGGUUGCAUGUGCAACCAAUUACAGUCGUUUCCGGC (((..............................((......))..........((((.(((.......))).((((((....))))))..........))))..))) (-15.92 = -15.80 + -0.12)

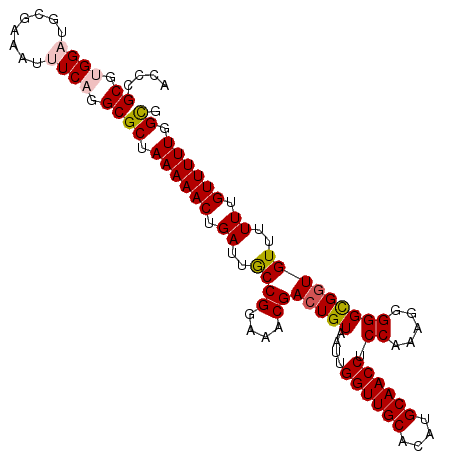
| Location | 17,617,789 – 17,617,895 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
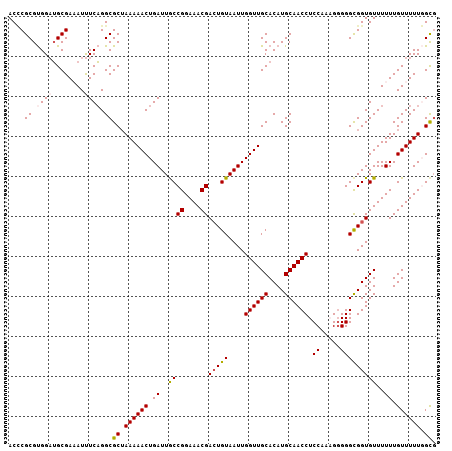
>2L_DroMel_CAF1 17617789 106 - 22407834 ACCCGCGUGGAUGCGAAAUUUCAGGCGCUAAAAACUGAUUGCCGGAAACGACUGUAAUUGGUUGCACAUGCAACCUCCAAAG-GGGCGGUGUUUUUUGUUUUUGGCG ....((.((((........)))).))(((((((((.((..((((....))(((((....((((((....)))))).((....-)))))))))..)).))))))))). ( -37.40) >DroSec_CAF1 13180 107 - 1 ACCCGCGUGGAUGCGAAAUCUCAGGCGCAAAAAACUGAUUACCGGAAACGACUGUAAUUGGUUGCACAUGCAACCUCCAAUGGGGGCGGUGUUUUUUGUUUUUGGCG ..((((.(((((.....))).)).))(((((((((.......((....))(((((....((((((....))))))(((....)))))))))))))))))....)).. ( -34.90) >DroSim_CAF1 3369 107 - 1 ACCCGCGUGGAUGCGAAAUUUCAGGCGCUAAAAACUGAUUGCCGGAAACGAAUGUAAUUGGUUGCACAUGCAACCUCCAAUGGGGGCGGUGUUUUUUGUUUUUGGCG ....((.((((........)))).))(((((((((..(((((((....))...)))))..)))((((.(((..((......))..)))))))........)))))). ( -35.80) >DroEre_CAF1 1599 105 - 1 ACUCGCGCGGUUGCGA-AUUUCAGGCGCUAAAAACUGAUUGCCGGAAACGACUGUAAUUGGUUGCACAUGCAACCUCCA-AGGGGGCGGUGUUUUUUGUUUUUCGCU ......((((..((((-(....(((((((...(((..(((((((....))...)))))..)))......((..((....-.))..))))))))))))))...)))). ( -31.20) >DroYak_CAF1 13130 106 - 1 ACGCGCGUGGAAGCGA-AUUUCUGGCGCUAAAAACUGAUUGCCGGAAACGACUGUAAUUGGUUGCACAUGCAACCUCCAAAGGGGGUGGUGUUUUUUGUUUUUCGUG ...((((.((((((((-(.....((((((...(((..(((((((....))...)))))..)))((....)).((((((...)))))))))))).))))))))))))) ( -34.10) >consensus ACCCGCGUGGAUGCGAAAUUUCAGGCGCUAAAAACUGAUUGCCGGAAACGACUGUAAUUGGUUGCACAUGCAACCUCCAAAGGGGGCGGUGUUUUUUGUUUUUGGCG ....((.((((........)))).))((.((((((.((..((((....))(((((....((((((....)))))).((.....)))))))))..)).)))))).)). (-27.18 = -27.70 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:17 2006