
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
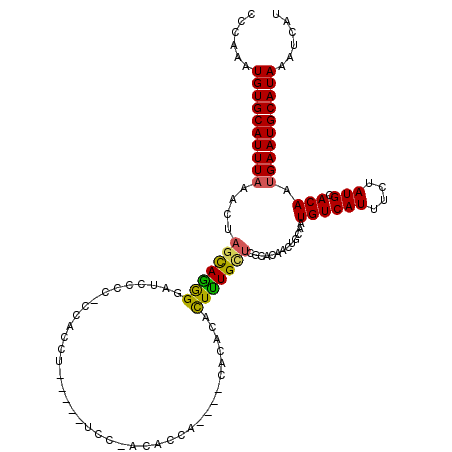
| Location | 17,617,543 – 17,617,648 |
| Length | 105 |
| Max. P | 0.998675 |

| Location | 17,617,543 – 17,617,648 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -15.31 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
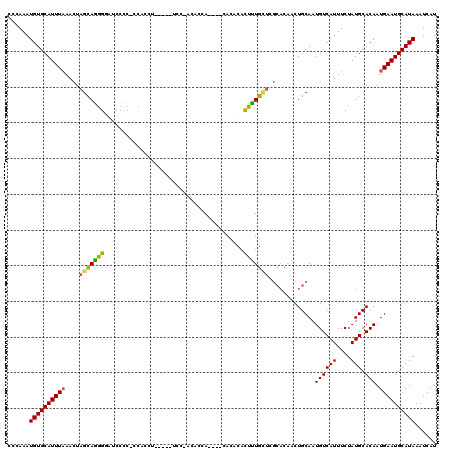
>2L_DroMel_CAF1 17617543 105 + 22407834 CCCAAAUGUGCAUUUAAACUAGCAAGGGAGCCACCCCAC-----------ACCACA----CACACACUUUGUUCGCACAACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((((.....(((.(((......)))..-----------......----........((((....)))).)))..((((((....))).))).))))))))))...... ( -20.50) >DroSim_CAF1 3112 116 + 1 CCCAAAUGUGCAUUUAAACUAGCAAGGGAUCCCCCCCACCUUUCAAUCCCACACCA----CACACACUUUGUUCGCACAACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((((.....(((.(((((...............)))))......----........((((....)))).)))..((((((....))).))).))))))))))...... ( -21.16) >DroEre_CAF1 1359 99 + 1 CCCAAAUGUGCAUUUAAACUAGCAGGGGAUCCCC--C---------------ACCA----CACACACUUUGCUCGCACAACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((((.....(((((((....))--)---------------....----.........((....))...))))..((((((....))).))).))))))))))...... ( -22.80) >DroYak_CAF1 12869 114 + 1 CCCAAAUGUGCAUUUAAACUAGCAGGGCAUCCCC--CACCUUUCGAUCCCACACCA----CACACACUUUGCUUGCACAACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((((.....((((((.(((...--........))))))......----..........(((((((....))))).))........)))....))))))))))...... ( -21.40) >DroPer_CAF1 4372 112 + 1 CCCAAAUGUGCAUUUCAAUUAGUAGAG-AGCCCCUCCCCCUGGAGCUCCUGC-CCAGGGGCACCCCUUCUGG------UACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((.((....((((..-........(((((((.((....))-))))))).(((......))------).))))..((((((....))).))).))))))))))...... ( -34.50) >consensus CCCAAAUGUGCAUUUAAACUAGCAGGGGAUCCCC_CCACCU_____UCC_ACACCA____CACACACUUUGCUCGCACAACUGCAAUGUCAUUUCUAUGCACAAUGAAUGCAUAAAUCAU ......((((((((((....(((((((.......................................))))))).............((((((....))).))).))))))))))...... (-15.31 = -14.87 + -0.44)

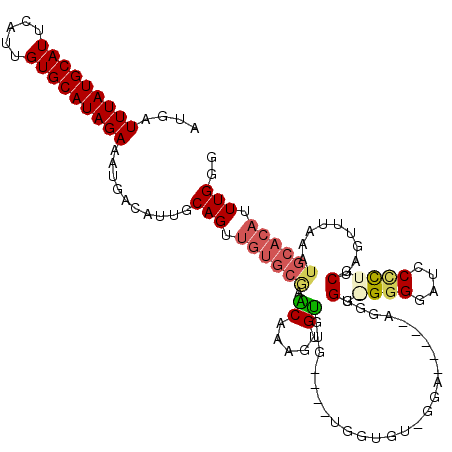
| Location | 17,617,543 – 17,617,648 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17617543 105 - 22407834 AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUUGUGCGAACAAAGUGUGUG----UGUGGU-----------GUGGGGUGGCUCCCUUGCUAGUUUAAAUGCACAUUUGGG ....(((((((((.....))))))))).......(((((....))))).(.(((((((((----(.(((.-----------(..(((......)))..)....))).)))))))))).). ( -28.20) >DroSim_CAF1 3112 116 - 1 AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUUGUGCGAACAAAGUGUGUG----UGGUGUGGGAUUGAAAGGUGGGGGGGAUCCCUUGCUAGUUUAAAUGCACAUUUGGG ....(((((((((.....)))))))))....((((.(((.((((....))))..))))))----)((((((.(.(((((.((..((((....))))..))..))))).).)))))).... ( -33.30) >DroEre_CAF1 1359 99 - 1 AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUUGUGCGAGCAAAGUGUGUG----UGGU---------------G--GGGGAUCCCCUGCUAGUUUAAAUGCACAUUUGGG ....(((((((((.....)))))))))..((...(((((....))))).))(((((((((----(((.---------------.--(((....)))..)).......))))))))))... ( -31.61) >DroYak_CAF1 12869 114 - 1 AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUUGUGCAAGCAAAGUGUGUG----UGGUGUGGGAUCGAAAGGUG--GGGGAUGCCCUGCUAGUUUAAAUGCACAUUUGGG ....(((((((((.....)))))))))..((...(((((....))))).))(((((((((----((((.....)))....((..--(((....)))..)).......))))))))))... ( -32.10) >DroPer_CAF1 4372 112 - 1 AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUA------CCAGAAGGGGUGCCCCUGG-GCAGGAGCUCCAGGGGGAGGGGCU-CUCUACUAAUUGAAAUGCACAUUUGGG ....(((((((((.....)))))))))...............------(((((....((((((((((-((....)).))))))((((.....-))))............)))).))))). ( -36.50) >consensus AUGAUUUAUGCAUUCAUUGUGCAUAGAAAUGACAUUGCAGUUGUGCGAACAAAGUGUGUG____UGGUGU_GGA_____AGGUGG_GGGGAUCCCCUGCUAGUUUAAAUGCACAUUUGGG ....(((((((((.....)))))))))..........(((.((((((.((.....))..........................(.((((....)))).).........)))))).))).. (-17.60 = -18.36 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:15 2006