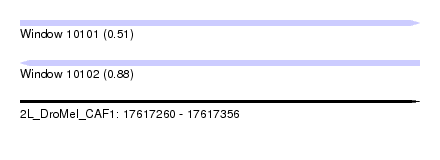
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,617,260 – 17,617,356 |
| Length | 96 |
| Max. P | 0.877803 |

| Location | 17,617,260 – 17,617,356 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17617260 96 + 22407834 AGUGAAAGAGCUGGGAGAACUCUUAAAAUUACUAACAGCUAACUAACAGUUAAAAG--AAAGAGAAUCGUGGCUAACAGAGCAUGCUCUUUGGGAAUU .((.....(((((..((..............))..))))).....)).........--((((((...((((.((.....))))))))))))....... ( -16.74) >DroSim_CAF1 2837 88 + 1 AGUGAAAGAGCUGGGAGAGCUCUUAAAAUAGCUAACAGCU--------GUUAAAAG--AAAGAGAAUCGUGGCUAACAGAGCAUGCUCUUUGGGAAUU ..........((.(((((((((((..((((((.....)))--------)))..)))--)............(((.....)))..))))))).)).... ( -22.30) >DroEre_CAF1 1089 90 + 1 AGUGAAAGAGCAGGAACAGCUCUUAAAACAGCUAACAGCG--------CUUAAGAGAGAGAGAGAAUCCUGGCCAACAGAGCAGGCUCUUUGGGAAAU ..........(((((....(((((......((.....)).--------(((....))).)))))..))))).((((.((((....))))))))..... ( -21.90) >DroYak_CAF1 12599 88 + 1 AGUGAAAGAGCAGCAAACGCUCUUAAAAUAGCUAACAGCG--------CUAAAAAG--UAAGAGAAUCGUGGCUAACAGAGCAGGCUCUUUGGGAAAU ..(.(((((((..((....((((((...((((.......)--------))).....--)))))).....))(((.....)))..))))))).)..... ( -21.70) >consensus AGUGAAAGAGCAGGAAGAGCUCUUAAAAUAGCUAACAGCG________CUUAAAAG__AAAGAGAAUCGUGGCUAACAGAGCAGGCUCUUUGGGAAAU ..(.(((((((........(((((...................................))))).......(((.....)))..))))))).)..... (-12.64 = -12.70 + 0.06)


| Location | 17,617,260 – 17,617,356 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -18.59 |
| Consensus MFE | -14.02 |
| Energy contribution | -13.65 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
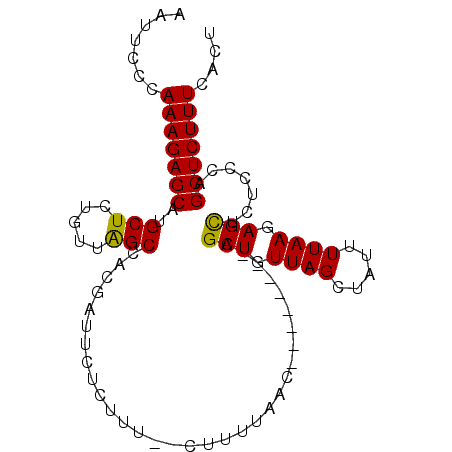
>2L_DroMel_CAF1 17617260 96 - 22407834 AAUUCCCAAAGAGCAUGCUCUGUUAGCCACGAUUCUCUUU--CUUUUAACUGUUAGUUAGCUGUUAGUAAUUUUAAGAGUUCUCCCAGCUCUUUCACU .......(((((((..(((.....)))...((..(((((.--...(((.(((.(((....))).))))))....))))).)).....))))))).... ( -18.50) >DroSim_CAF1 2837 88 - 1 AAUUCCCAAAGAGCAUGCUCUGUUAGCCACGAUUCUCUUU--CUUUUAAC--------AGCUGUUAGCUAUUUUAAGAGCUCUCCCAGCUCUUUCACU .......(((((((..(((((..((((...((.......)--)...((((--------....)))))))).....))))).......))))))).... ( -17.60) >DroEre_CAF1 1089 90 - 1 AUUUCCCAAAGAGCCUGCUCUGUUGGCCAGGAUUCUCUCUCUCUCUUAAG--------CGCUGUUAGCUGUUUUAAGAGCUGUUCCUGCUCUUUCACU .....((((((((....)))).)))).(((((....((((........((--------(.......)))......))))....))))).......... ( -20.04) >DroYak_CAF1 12599 88 - 1 AUUUCCCAAAGAGCCUGCUCUGUUAGCCACGAUUCUCUUA--CUUUUUAG--------CGCUGUUAGCUAUUUUAAGAGCGUUUGCUGCUCUUUCACU .......(((((((..((...((.....))(((.((((((--.....(((--------(.......))))...)))))).))).)).))))))).... ( -18.20) >consensus AAUUCCCAAAGAGCAUGCUCUGUUAGCCACGAUUCUCUUU__CUUUUAAC________AGCUGUUAGCUAUUUUAAGAGCUCUCCCAGCUCUUUCACU .......(((((((..(((.....)))................................(((.((((.....)))).))).......))))))).... (-14.02 = -13.65 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:13 2006