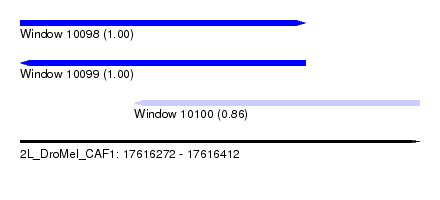
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,616,272 – 17,616,412 |
| Length | 140 |
| Max. P | 0.999967 |

| Location | 17,616,272 – 17,616,372 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17616272 100 + 22407834 CCACUAUCGCUGGCCGUAUCUUGCUGCCAAAUUCGCUGAUUCACGUAAUUUGCCGCGACUGCAGCAGCGACAACGGCCAAAAGCCACGGCCAACAACCAA ........(.(((((((...(((((((((((((((........)).))))))..((....)).)))))))....(((.....)))))))))).)...... ( -31.80) >DroSec_CAF1 11598 100 + 1 CCACUAUCGCUGGCCGUAUCUUUCUGCCAAAUUCGCUGAUUCACGUAAUUUGCCGCGACUGCAGCAGCGACAACGGCCAAAAGCCACGGCCAAAAACCAA ..........(((((((..((((..(((....((((((...(((((........)))..))...))))))....)))..))))..)))))))........ ( -31.10) >DroSim_CAF1 1840 100 + 1 CCACUAUCGCUGGCCGUAUCUUGCUGCCAAAUUCGCUGAUUCACGUAAUUUGCCGCGACUGCAGCAGCGACAACGGCCAAAAGCCACGGCCAACAACCAA ........(.(((((((...(((((((((((((((........)).))))))..((....)).)))))))....(((.....)))))))))).)...... ( -31.80) >DroEre_CAF1 80 100 + 1 CCUCUAUCGUUGGCCGUAUCUUGCUGCCAAAUUCGCUGAUUCACGUAAUUUCCCGCGACUGCAGCAGCGACAACGGCCAAAAGCCAUGGCCAACAACCAA ........(((((((((...(((((((.....((((.((...........))..)))).....)))))))....(((.....))))))))))))...... ( -33.50) >DroYak_CAF1 11547 100 + 1 CCACUAUCGUUGGCCGUAUCUUGCUGCCAAAUUCGCUGAUUCACGUAAUUUCCCGCGACUGCAGCACCGACAACGGCCAAAAGCCAUGGCCAGCAACCAA ........(((((((((....((((((.....((((.((...........))..))))..))))))........(((.....))))))))))))...... ( -31.80) >consensus CCACUAUCGCUGGCCGUAUCUUGCUGCCAAAUUCGCUGAUUCACGUAAUUUGCCGCGACUGCAGCAGCGACAACGGCCAAAAGCCACGGCCAACAACCAA ..........(((((((..(((...(((....((((((...(((((........)))..))...))))))....)))...)))..)))))))........ (-28.24 = -28.20 + -0.04)


| Location | 17,616,272 – 17,616,372 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -43.54 |
| Consensus MFE | -41.08 |
| Energy contribution | -41.96 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.99 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17616272 100 - 22407834 UUGGUUGUUGGCCGUGGCUUUUGGCCGUUGUCGCUGCUGCAGUCGCGGCAAAUUACGUGAAUCAGCGAAUUUGGCAGCAAGAUACGGCCAGCGAUAGUGG ...(((((((((((((.(((...((((...((((((......(((((........)))))..))))))...))))...))).)))))))))))))..... ( -46.30) >DroSec_CAF1 11598 100 - 1 UUGGUUUUUGGCCGUGGCUUUUGGCCGUUGUCGCUGCUGCAGUCGCGGCAAAUUACGUGAAUCAGCGAAUUUGGCAGAAAGAUACGGCCAGCGAUAGUGG ...(((.(((((((((.(((((.((((...((((((......(((((........)))))..))))))...)))).))))).))))))))).)))..... ( -43.40) >DroSim_CAF1 1840 100 - 1 UUGGUUGUUGGCCGUGGCUUUUGGCCGUUGUCGCUGCUGCAGUCGCGGCAAAUUACGUGAAUCAGCGAAUUUGGCAGCAAGAUACGGCCAGCGAUAGUGG ...(((((((((((((.(((...((((...((((((......(((((........)))))..))))))...))))...))).)))))))))))))..... ( -46.30) >DroEre_CAF1 80 100 - 1 UUGGUUGUUGGCCAUGGCUUUUGGCCGUUGUCGCUGCUGCAGUCGCGGGAAAUUACGUGAAUCAGCGAAUUUGGCAGCAAGAUACGGCCAACGAUAGAGG ...((((((((((..(((.....)))(((((((.(((((...(((((........)))))..)))))....))))))).......))))))))))..... ( -42.40) >DroYak_CAF1 11547 100 - 1 UUGGUUGCUGGCCAUGGCUUUUGGCCGUUGUCGGUGCUGCAGUCGCGGGAAAUUACGUGAAUCAGCGAAUUUGGCAGCAAGAUACGGCCAACGAUAGUGG ...((((.(((((..(((.....)))(((((((.(((((...(((((........)))))..)))))....))))))).......))))).))))..... ( -39.30) >consensus UUGGUUGUUGGCCGUGGCUUUUGGCCGUUGUCGCUGCUGCAGUCGCGGCAAAUUACGUGAAUCAGCGAAUUUGGCAGCAAGAUACGGCCAGCGAUAGUGG ...(((((((((((((.(((...((((...((((((......(((((........)))))..))))))...))))...))).)))))))))))))..... (-41.08 = -41.96 + 0.88)



| Location | 17,616,312 – 17,616,412 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -20.77 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17616312 100 - 22407834 UAGUGGCGGGAGAGACAACACAUUGGCUACGGCUUUGGCUUUGGUUGUUGGCCGUGG-----CUUUUGGCCGUUGUCGCUG------CUGCAGUCGCGGCAAAUUACGUGA .....((((.((.((((((.((..((((((((((.((((....))))..))))))))-----))..))...)))))).)).------))))..(((((........))))) ( -44.70) >DroPse_CAF1 3281 103 - 1 UAAUGGGG-CAGUGGCAUGACAUUG-------CUUUGGCUUUGGGCCUGGACCAUGAUGUCGGUUGUCGGUGCCGUCGCUGUUGUGCCUGCAGUCGCGGCAAAUUACGUGA ..(((.((-((.((((((((((((.-------((..(((.....))).)).....)))))))..))))).))))(((((..((((....))))..)))))......))).. ( -36.40) >DroSec_CAF1 11638 100 - 1 UAGUGGCGGGUGAGACAACACAUUGGCUACGGCUUUGGCUUUGGUUUUUGGCCGUGG-----CUUUUGGCCGUUGUCGCUG------CUGCAGUCGCGGCAAAUUACGUGA ..(..((((....((((((.((..((((((((((..(((....)))...))))))))-----))..))...)))))).)))------)..)..(((((........))))) ( -44.60) >DroSim_CAF1 1880 100 - 1 UAGUGGCGGGUGAGACAACACAUUGGCUACGGCUUUGGCUUUGGUUGUUGGCCGUGG-----CUUUUGGCCGUUGUCGCUG------CUGCAGUCGCGGCAAAUUACGUGA ..(..((((....((((((.((..((((((((((.((((....))))..))))))))-----))..))...)))))).)))------)..)..(((((........))))) ( -43.90) >DroEre_CAF1 120 99 - 1 UAAUGGCCGGA-CGACAAGCCAUUGGCUAUGGUAUUGGCUUUGGUUGUUGGCCAUGG-----CUUUUGGCCGUUGUCGCUG------CUGCAGUCGCGGGAAAUUACGUGA ....(((.(((-((((..((((..(((((((((..((((....))))...)))))))-----))..)))).)))))).).)------))....(((((........))))) ( -40.00) >DroYak_CAF1 11587 99 - 1 UAUUUGCGGGA-CGACAACACAUUGGCUAUGGUAUUGGCUUUGGUUGCUGGCCAUGG-----CUUUUGGCCGUUGUCGGUG------CUGCAGUCGCGGGAAAUUACGUGA ...((((((.(-(((((((.((..(((((((((...(((.......))).)))))))-----))..))...)))))).)).------))))))(((((........))))) ( -42.60) >consensus UAGUGGCGGGAGAGACAACACAUUGGCUACGGCUUUGGCUUUGGUUGUUGGCCAUGG_____CUUUUGGCCGUUGUCGCUG______CUGCAGUCGCGGCAAAUUACGUGA .............((((((.....(((((.(((....))).)))))...(((((..(......)..)))))))))))................(((((........))))) (-20.77 = -21.75 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:11 2006