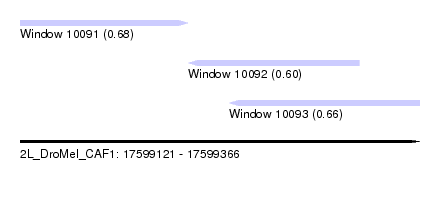
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,599,121 – 17,599,366 |
| Length | 245 |
| Max. P | 0.679899 |

| Location | 17,599,121 – 17,599,224 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -15.12 |
| Energy contribution | -16.29 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
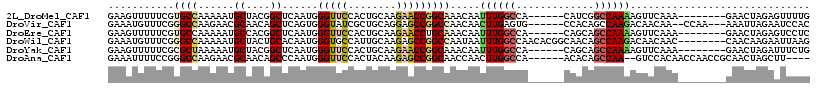
>2L_DroMel_CAF1 17599121 103 + 22407834 GAAGUUUUUCGUGCCAAAAAUGCUACGGCUCAAUGGGUUCCACUGCAAGAACCGGCAAACAAUUUGGCCA------CAUCGGCCAAAAGUUCAAA--------GAACUAGAGUUUUG ..(((((((..((((......((....))......(((((........))))))))).....(((((((.------....)))))))......))--------)))))......... ( -30.10) >DroVir_CAF1 2509 106 + 1 GAAAUGUUUCGGGCCAAGAACGCAACAGCUCAGUGGGUAUCGCUGCAGGAGCCGGCCAACAACUUGAGUG------CCACAGCCAAGACAACAA--CCAA---AAAUUAGAAUCCAC ((..((((.((..(...)..)).))))..)).((((((.(((((((....).))))......((((.((.------.....)))))).......--....---......)))))))) ( -20.50) >DroEre_CAF1 2665 103 + 1 GAAGUUUUUCGUGCCAAAAAUGCCACGGCUCAAUGGGUUCCACUGCAAGAACCUGCAAACAAUUUGGCCA------CAGCAGCCAAAAGUUCAAA--------GAACUAGAGUCCUC ..........(((.((....)).)))(((((...((((((........))))))........((((((..------.....))))))(((((...--------))))).)))))... ( -24.60) >DroWil_CAF1 1371 109 + 1 GAAAUGUUUCGGGCCAAAAAUGCUACUGCACAAUGGGUGCCAUUGCAAGAGCCGGCCAAUAAUUUGGCCAACACGGCAACAGCCAAGACAACAAC--------CAACAAGAAUUAAG ....((((((.((((.....(((....)))((((((...))))))...).)))((((((....)))))).....(((....)))..)).))))..--------.............. ( -29.40) >DroYak_CAF1 2283 103 + 1 GAAGUUUUUCGCGCUAAAAAUGCUACGGCUCAAUGGGUUCCACUGCAAGAACCGGCAAACAAUUUGGCCA------CAGCAGCCAAAAGUUCAAA--------GAACUAGAUUUCUG (((((((.....(((......((....))......(((((........))))))))......((((((..------.....))))))(((((...--------)))))))))))).. ( -24.50) >DroAna_CAF1 2065 105 + 1 GAAAUUUUCCGGGCCAAGAACGCAACAGCCCAAUGGGUUCCACUACAAGAGCCGGCAACCAACUUGGCCA------ACACAGCCAA--GUCCACAACCAACCGCAACUAGCUU---- .........((((((......((....))......(((((........)))))))).....(((((((..------.....)))))--))..........)))..........---- ( -24.70) >consensus GAAAUUUUUCGGGCCAAAAAUGCUACGGCUCAAUGGGUUCCACUGCAAGAACCGGCAAACAAUUUGGCCA______CAACAGCCAAAAGUUCAAA________GAACUAGAAUUCUG ...........((((......((....))......(((((........))))))))).....((((((.............)))))).............................. (-15.12 = -16.29 + 1.17)
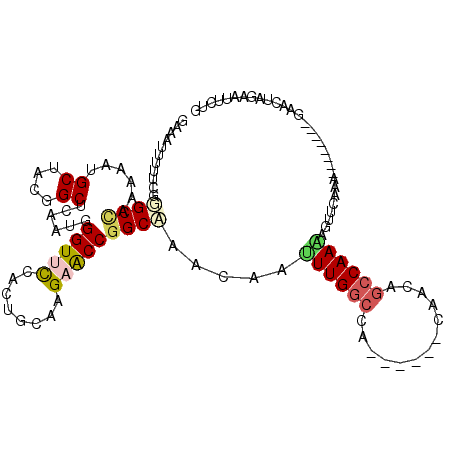
| Location | 17,599,224 – 17,599,329 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.12 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -11.74 |
| Energy contribution | -14.22 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17599224 105 - 22407834 GGAUAUUUUGGUAUUUUUUUAUCGCCUGCAACCGAGUACACACACAUCCUUAGAAAUAAGGAUGACAUCUGUGGAAAUACUGAAAAAAAUACCUUUACAAUAAUA .........(((((((((((.(((........)))(((..((((((((((((....)))))))).....))))....)))..)))))))))))............ ( -25.90) >DroSec_CAF1 4443 103 - 1 GGAUAUUUCGGUAUUU-UUUAUCGCCAGGAAACCAGUACACACACAUCGUUGGAAACUAGGAUGGCAUCUGUGGAAAUACUGGAA-AAAUACCUUUACAAAAAUA .........(((((((-(((.......(....)(((((..((((((((.(.(....).).)))).....))))....))))))))-)))))))............ ( -25.00) >DroSim_CAF1 4374 103 - 1 GGAUAUUUCGGUAUUU-UUUAUCGCCAGGAACCCAGUACACACACAUCCUUGGAAACUAGGAUGGCAUCUGUGGAAAUACUGGAA-AAAUACCUUUACAAAAAUA .........(((((((-((..((.....))..((((((..((((((((((.(....).)))))).....))))....))))))))-)))))))............ ( -28.80) >DroEre_CAF1 2768 92 - 1 ----------GUACU--UCAAUCGGCUAGAACCGACUGCACAUACAUCCUUAGAAACUAGGAUGGCAUCUGAGGAAAUACUGGAG-AAAUACCUUUACAAUAUUA ----------...((--(((.((((......))))...........((((((((..((.....))..)))))))).....)))))-................... ( -18.80) >DroYak_CAF1 2386 102 - 1 GGAUAUUUUGGUAUU--UUAAUCGACUAGAACCGAGUACACACACAUCCUUUGAAACUAGGAUGGCAUCGGUGGAAAUACUGGAA-AAAUACUUUUACAAUAAUG .....(((..(((((--((..((.....))(((((((....)).((((((........))))))...))))).)))))))..)))-................... ( -23.20) >consensus GGAUAUUUCGGUAUUU_UUUAUCGCCUGGAACCGAGUACACACACAUCCUUAGAAACUAGGAUGGCAUCUGUGGAAAUACUGGAA_AAAUACCUUUACAAUAAUA .........(((((((((((.(((........)))...((((..((((((.(....).)))))).....)))).........)))))))))))............ (-11.74 = -14.22 + 2.48)


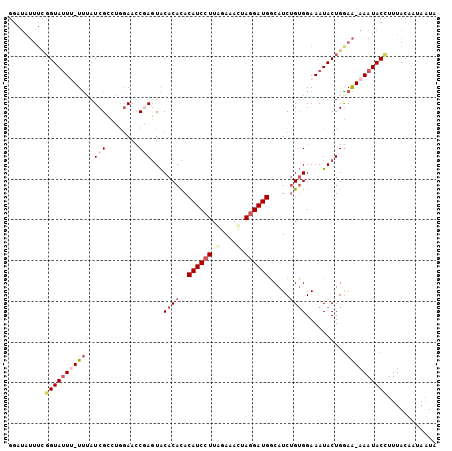
| Location | 17,599,249 – 17,599,366 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -15.40 |
| Energy contribution | -17.56 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
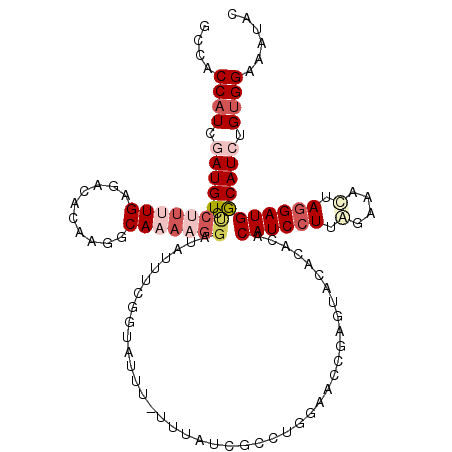
>2L_DroMel_CAF1 17599249 117 - 22407834 GCCACCAUCGAUGUCUCUUUUGAGACACAAGGCAAAAGGAUAUUUUGGUAUUUUUUUAUCGCCUGCAACCGAGUACACACACAUCCUUAGAAAUAAGGAUGACAUCUGUGGAAAUAC .((((..(((.((((((....))))))..((((((((((((((....))))))))))...)))).....))).........((((((((....))))))))......))))...... ( -37.00) >DroSec_CAF1 4467 115 - 1 GCCACCAUCGAUGUUCCUUUUGAGA-ACACGGCAAACGGAUAUUUCGGUAUUU-UUUAUCGCCAGGAAACCAGUACACACACAUCGUUGGAAACUAGGAUGGCAUCUGUGGAAAUAC ....((((.(((((((((..((...-.)).(((.....((....))((((...-..)))))))))))..............((((.(.(....).).))))))))).))))...... ( -26.70) >DroSim_CAF1 4398 116 - 1 GCCACCAUCGAUGUCUCCUUUGAGACACAAGGCAAACGGAUAUUUCGGUAUUU-UUUAUCGCCAGGAACCCAGUACACACACAUCCUUGGAAACUAGGAUGGCAUCUGUGGAAAUAC ....((((.(((((.(((((((.....)))(((.....((....))((((...-..)))))))))))..............((((((.(....).))))))))))).))))...... ( -32.10) >DroEre_CAF1 2792 105 - 1 GCAACCAUGAAUGUAUCUUAUGAGACACAAUCCAAAA----------GUACU--UCAAUCGGCUAGAACCGACUGCACAUACAUCCUUAGAAACUAGGAUGGCAUCUGAGGAAAUAC (((....((((.(((.(((.((.((.....)))).))----------)))))--))).((((......)))).))).......((((((((..((.....))..))))))))..... ( -23.40) >DroYak_CAF1 2410 115 - 1 GCAACCAUUAAUGUCUCUUAUGAGACACAAUCCAACAGGAUAUUUUGGUAUU--UUAAUCGACUAGAACCGAGUACACACACAUCCUUUGAAACUAGGAUGGCAUCGGUGGAAAUAC ...((((....((((((....))))))..((((....))))....))))(((--((..((.....))(((((((....)).((((((........))))))...))))).))))).. ( -28.70) >consensus GCCACCAUCGAUGUCUCUUUUGAGACACAAGGCAAAAGGAUAUUUCGGUAUUU_UUUAUCGCCUGGAACCGAGUACACACACAUCCUUAGAAACUAGGAUGGCAUCUGUGGAAAUAC ....((((.(((((.(((((((..........)))))))..........................................((((((.(....).))))))))))).))))...... (-15.40 = -17.56 + 2.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:05 2006