
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,590,161 – 17,590,270 |
| Length | 109 |
| Max. P | 0.722189 |

| Location | 17,590,161 – 17,590,270 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.30 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.11 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17590161 109 - 22407834 ACUACCGGAAGGAAAAGCCUUUUUUUAUACAAAAGCUUCCCAAAGAGCAACAGCUGGCCAAUUUUAGAAAAAAGCCCGCGAAAUCCGAAUCGGUUAA--------CCUUUCCCAA-AA ......((((((...((((...............((((......))))....((.(((...............))).))............))))..--------.))))))...-.. ( -20.16) >DroPse_CAF1 114168 101 - 1 CUAAUCGGC-GGCAAAUCCC-C-CCAUCUUGGAAGCUCUCCGGAAAGCAACAGCUGGCACAUUUGG--A-AAAGCCCGCGCAACGG-------GGAGUGCGUUGCC-U-GGAUAAA-- ......(((-(((.......-.-((.....))..((((((((....((....((.(((........--.-...))).))))..)))-------)))))..))))))-.-.......-- ( -30.10) >DroSec_CAF1 107000 107 - 1 ACUACCGGAAGGAAAAGCCUUU-UUUACACAAAAGCUUCCCAAAGAGCAACAGCUGGCCAAUUUUAUAA-AAAGCCCGCGAAAUCCGAAUCGGUUAA--------CCUUUUCCAA-AA ......((((((...((((...-...........((((......))))....((.(((...........-...))).))............))))..--------)))..)))..-.. ( -18.84) >DroSim_CAF1 106027 107 - 1 ACUACCGGAAGGAAAGGCCUUU-UUUACACAAAAGCUUCCCAAAGAGCAACAGCUGGCCCAUUUUAUAA-AAAGCCCGCGAAAUCCGAAUCGGUUAA--------CCUUUUCCAA-AA ......((((((...((((...-...........((((......))))....((.(((...........-...))).))............))))..--------)))..)))..-.. ( -18.74) >DroEre_CAF1 105108 113 - 1 ACUACCGGAAGGAAAAGCCU-U-CUUACACAAAAGCUCCCCAAAGAGCAACAGCUGGCCAAUUUUGUAA-AAAGCCCGCGAAAUUCCAAACGGUUAAUACUUAGACCUUUCCAAAA-- ......((((((((......-.-...........((((......))))....((.(((...........-...))).))....))))....((((........)))).))))....-- ( -23.34) >DroYak_CAF1 107827 116 - 1 AAUACCGGAAGGAAAAGCCUUU-CUUACACAAAAGCUCCUCACAGAGCAACAGCUGGCCAAUUUCAUAA-AAAGCCCGCGAAAUCCCAAGCGGUUAAUAUUUAGACCUUUCCCAAAAA ......((((((.....)))))-)..........((((......))))....((.(((...........-...))).))............((((........))))........... ( -20.64) >consensus ACUACCGGAAGGAAAAGCCUUU_CUUACACAAAAGCUCCCCAAAGAGCAACAGCUGGCCAAUUUUAUAA_AAAGCCCGCGAAAUCCGAAUCGGUUAA________CCUUUCCCAA_AA ....((....))...((((...............((((......))))....((.(((...............))).))............))))....................... (-15.14 = -15.11 + -0.03)

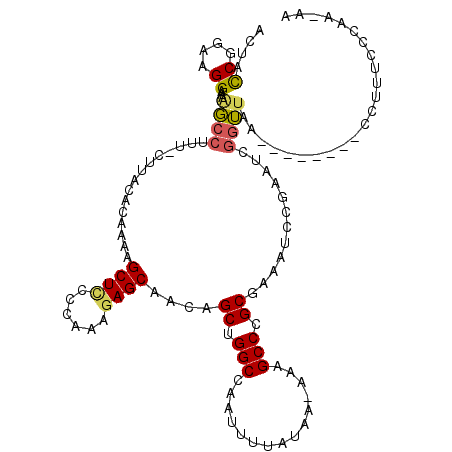
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:02 2006