| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,588,426 – 17,588,562 |
| Length | 136 |
| Max. P | 0.920247 |

| Location | 17,588,426 – 17,588,524 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -10.73 |
| Energy contribution | -12.26 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
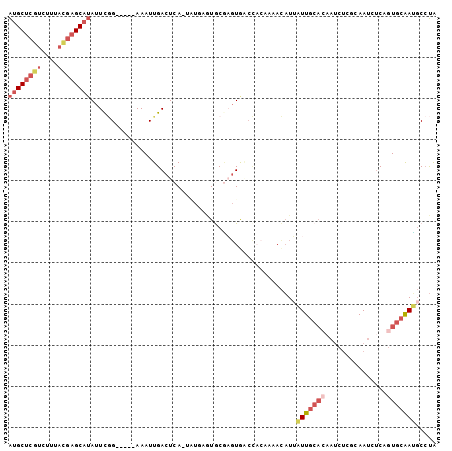
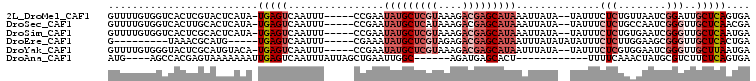
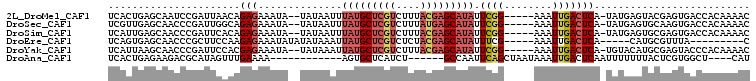
>2L_DroMel_CAF1 17588426 98 - 22407834 AUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUACGAGUGACCACAAAACAUUAUUGCAAAAUCUCGCAAUCUCAGUGCAAUGCCUA ((((((((....))))))))....((-----...(((((((.-...)))).)))....))......(((((((((........)))))...))))......... ( -21.70) >DroSec_CAF1 105182 98 - 1 AUGCUCGUCUUUAUGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUGCAAGUGACCACAAAACAUUAUUGCACAAUCUCGCAAUCUCAGUGCAAUGCCUA ((((((((....))))))))....((-----...(((((((.-...)))).)))....)).........((((((((...............)))))))).... ( -23.56) >DroSim_CAF1 104268 98 - 1 AUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUGCGAGUGACCACAAAACAUUAUUGCACAAUCUCGCAAUCUCAGUGCAAUGCCUA ((((((((....))))))))....((-----...(((((((.-...)))).)))....)).........((((((((...............)))))))).... ( -25.06) >DroEre_CAF1 103392 85 - 1 AUGCUCGUCUCUACGAGCAUAUUUCG-----AAAUUGACUCA-----CAUGCGUUUA---------CAUAGUUGCACAAUCUCCCAAUCCCAGUGCAACGCCUA ((((((((....))))))))......-----...........-----..........---------....(((((((...............)))))))..... ( -19.66) >DroYak_CAF1 106078 98 - 1 AUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UGUACAUGCGAGUACCCACAAAACAUUAUUGCACAAUCUCUUAAUCUCAGUGCAAUGCCUA ((((((((....))))))))....((-----...........-.((((......))))...........((((((((...............)))))))))).. ( -21.96) >DroAna_CAF1 109670 86 - 1 GUGCUCAUCU------GCCAAUUCAGCUAAUAAAUUGACUCAAUUUUUUUACUCGUGGCU----CAUUUUAUU--------ACCUAAUCUCGGUGCGAAUCACG (((.((....------((((....((..((.((((((...)))))).))..))..)))).----........(--------(((.......)))).))..))). ( -12.40) >consensus AUGCUCGUCUUUACGAGCAUAUUCGG_____AAAUUGACUCA_UAUGAGUGCGAGUGACCACAAAACAUUAUUGCACAAUCUCGCAAUCUCAGUGCAAUGCCUA ((((((((....))))))))..................................................(((((((...............)))))))..... (-10.73 = -12.26 + 1.53)


| Location | 17,588,463 – 17,588,562 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.15 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -9.79 |
| Energy contribution | -11.65 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17588463 99 + 22407834 GUUUUGUGGUCACUCGUACUCAUA-UGAGUCAAUUU-----CCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUA--UAUUUCUCUGUUAAUCGGAUUGCUCAGUGA ...................((((.-((((.(((..(-----((((.(((((((((....)))))))))(((....--.)))..........)))))))))))))))) ( -23.90) >DroSec_CAF1 105219 99 + 1 GUUUUGUGGUCACUUGCACUCAUA-UGAGUCAAUUU-----CCGAAUAUGCUCAUAAAGACGAGCAUAAAUUAUA--UAUUUCUCUGCCAAUCGGGUUGCUCAACGA (((..(..(....(((.((((...-.)))))))..(-----((((.(((((((........)))))))(((....--.)))..........))))))..)..))).. ( -19.80) >DroSim_CAF1 104305 99 + 1 GUUUUGUGGUCACUCGCACUCAUA-UGAGUCAAUUU-----CCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUA--UAUUUCUCUGUGAAUCGGGUUGCUCAAUGA ....(((((....))))).((((.-((((.((((..-----((((.(((((((((....)))))))))..(((((--........))))).)))))))))))))))) ( -28.70) >DroEre_CAF1 103429 88 + 1 G---------UAAACGCAUG-----UGAGUCAAUUU-----CGAAAUAUGCUCGUAGAGACGAGCAUAAUUUAUAUAUAUUUCUCUUGGAAGCGGGUUGCUCACUGA .---------......((.(-----((((.(((((.-----((...(((((((((....)))))))))...........((((....)))).)))))))))))))). ( -25.10) >DroYak_CAF1 106115 99 + 1 GUUUUGUGGGUACUCGCAUGUACA-UGAGUCAAUUU-----CCGAAUAUGCUCGUAAAGACGAGCAUAAUUUAUA--UAUUUCUCGUGGAAUCGGGUUGCUUAAUGA ......((((((((((.((...((-((((.......-----.....(((((((((....))))))))).......--.....))))))..)))))).)))))).... ( -28.85) >DroAna_CAF1 109699 85 + 1 AUG----AGCCACGAGUAAAAAAAUUGAGUCAAUUUAUUAGCUGAAUUGGC------AGAUGAGCACU------------UUUUCAAACUAUGCGUCUUCUCAGUGA .((----((..(((.(((..........(((((((((.....)))))))))------...((((....------------..))))...))).)))...)))).... ( -14.60) >consensus GUUUUGUGGUCACUCGCACUCAUA_UGAGUCAAUUU_____CCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUA__UAUUUCUCUGGGAAUCGGGUUGCUCAAUGA .........................(((((................(((((((((....)))))))))..............(((........)))..))))).... ( -9.79 = -11.65 + 1.86)
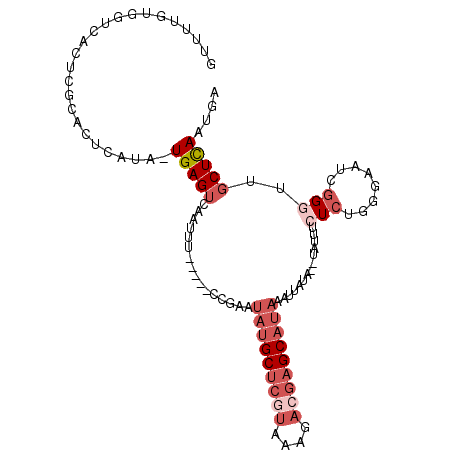

| Location | 17,588,463 – 17,588,562 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -9.09 |
| Energy contribution | -10.23 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17588463 99 - 22407834 UCACUGAGCAAUCCGAUUAACAGAGAAAUA--UAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUACGAGUGACCACAAAAC (((((......(((((.......(....).--.......(((((((((....))))))))).))))-----).....((((.-...))))...)))))......... ( -24.90) >DroSec_CAF1 105219 99 - 1 UCGUUGAGCAACCCGAUUGGCAGAGAAAUA--UAUAAUUUAUGCUCGUCUUUAUGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUGCAAGUGACCACAAAAC (((((((((((.((((.......(....).--.......(((((((((....))))))))).))))-----...))).))))-.))))(((........)))..... ( -21.50) >DroSim_CAF1 104305 99 - 1 UCAUUGAGCAACCCGAUUCACAGAGAAAUA--UAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UAUGAGUGCGAGUGACCACAAAAC (((((..((...(((((((.....)))...--.......(((((((((....))))))))).))))-----......((((.-...)))))).)))))......... ( -25.60) >DroEre_CAF1 103429 88 - 1 UCAGUGAGCAACCCGCUUCCAAGAGAAAUAUAUAUAAAUUAUGCUCGUCUCUACGAGCAUAUUUCG-----AAAUUGACUCA-----CAUGCGUUUA---------C ...((((((((....(((....)))..............(((((((((....))))))))).....-----...))).))))-----).........---------. ( -20.00) >DroYak_CAF1 106115 99 - 1 UCAUUAAGCAACCCGAUUCCACGAGAAAUA--UAUAAAUUAUGCUCGUCUUUACGAGCAUAUUCGG-----AAAUUGACUCA-UGUACAUGCGAGUACCCACAAAAC .......(((..(((((((.....)))...--.......(((((((((....))))))))).))))-----....((((...-.)).)))))............... ( -19.20) >DroAna_CAF1 109699 85 - 1 UCACUGAGAAGACGCAUAGUUUGAAAA------------AGUGCUCAUCU------GCCAAUUCAGCUAAUAAAUUGACUCAAUUUUUUUACUCGUGGCU----CAU ......(((.((.((((...((....)------------))))))).)))------((((....((..((.((((((...)))))).))..))..)))).----... ( -13.00) >consensus UCAUUGAGCAACCCGAUUCACAGAGAAAUA__UAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGG_____AAAUUGACUCA_UAUGAGUGCGAGUGACCACAAAAC ......................(((..............(((((((((....))))))))).((((........))))))).......................... ( -9.09 = -10.23 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:01 2006