| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,584,121 – 17,584,256 |
| Length | 135 |
| Max. P | 0.979399 |

| Location | 17,584,121 – 17,584,228 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
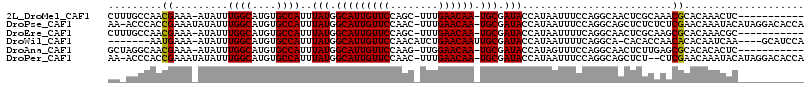
>2L_DroMel_CAF1 17584121 107 + 22407834 CUAUCAUUUUCCAUA---CCUUUUUCUAGCUUAUG--CCUUUGCCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAGCUUUGAACAAUGCGAUACCAUAAUU .((((..........---..........((.((((--((.............-......)))))).))........(((((((((.......)))))))))))))........ ( -23.01) >DroPse_CAF1 106862 109 + 1 CUCUCAUGGGCAAUA---CCCCUUCAAAGCGUGUGAGAAA-ACCCACCGAAAUAUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAACUUUGAACAAUGCGAUACCAUAAUU .(((((((.((....---..........)).)))))))..-...........((((..((((....))))..))))(((((((((.......)))))))))............ ( -29.64) >DroEre_CAF1 99213 107 + 1 CGAUCAUUUUCCAUG---CCUUUCUCCAGCGUAUG--CCUUUGCCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAGCUUUGAACAAUGCGAUACCAUAAUU ...........((((---((.........(((..(--(....))..)))...-......)))))).......(((.(((((((((.......))))))))).)))........ ( -25.67) >DroYak_CAF1 101843 106 + 1 CGAUCAUUUUCCAUA---CCUUUUUCUAGCGUAUG--CCU-UGCCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAGCUUUGAACAAUGCGAUACCAUAAUU ..(((......((((---(((......)).)))))--...-...........-(((..((((....))))..))).(((((((((.......))))))))))))......... ( -24.50) >DroAna_CAF1 105625 108 + 1 CUAG--AUUUCAAAAGGUCCUUCUUCUAGGGUAUG--CGCUAGGCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAAGUUGGAACAAUGCGAUACCAUAGUU ((((--(.................)))))(((((.--(....(....)....-(((..((((....))))..))).((((((((((.....))))))))))))))))...... ( -33.23) >DroPer_CAF1 107298 109 + 1 CUCUCAUGGGCAAUA---CCCCUUCAAAGUGUGUGAGAAA-ACCCACCGAAAUAUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAACUUUGAACAAUGCGAUACCAUAAUU .(((((((.((....---..........)).)))))))..-...........((((..((((....))))..))))(((((((((.......)))))))))............ ( -28.14) >consensus CUAUCAUUUUCAAUA___CCUUUUUCUAGCGUAUG__CCU_AGCCAACGAAA_AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAACUUUGAACAAUGCGAUACCAUAAUU ...............................((((.......................((((....))))..(((.(((((((((.......))))))))).))).))))... (-20.54 = -20.32 + -0.22)


| Location | 17,584,121 – 17,584,228 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17584121 107 - 22407834 AAUUAUGGUAUCGCAUUGUUCAAAGCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGGCAAAGG--CAUAAGCUAGAAAAAGG---UAUGGAAAAUGAUAG ........(((((((((((((.......)))))))))((((..((((....))))....(-(((((((.((....)--)...)))..)))))...---))))......)))). ( -25.70) >DroPse_CAF1 106862 109 - 1 AAUUAUGGUAUCGCAUUGUUCAAAGUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAUAUUUCGGUGGGU-UUUCUCACACGCUUUGAAGGGG---UAUUGCCCAUGAGAG ..(((((.(((.(((((((((.......))))))))).)))...((((.(((((..........(((((((.-...))))).)).........))---))))))))))))... ( -33.21) >DroEre_CAF1 99213 107 - 1 AAUUAUGGUAUCGCAUUGUUCAAAGCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGGCAAAGG--CAUACGCUGGAGAAAGG---CAUGGAAAAUGAUCG ..(((((.....(((((((((.......)))))))))))))).((((....))))..(((-((((((..((....)--)..))(((.......))---)...))))))).... ( -26.90) >DroYak_CAF1 101843 106 - 1 AAUUAUGGUAUCGCAUUGUUCAAAGCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGGCA-AGG--CAUACGCUAGAAAAAGG---UAUGGAAAAUGAUCG ..(((((.....(((((((((.......)))))))))))))).((((....))))..(((-(((((....).-...--(((((.((......)))---))))))))))).... ( -26.60) >DroAna_CAF1 105625 108 - 1 AACUAUGGUAUCGCAUUGUUCCAACUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGCCUAGCG--CAUACCCUAGAAGAAGGACCUUUUGAAAU--CUAG ..(((.((((.(((((((((((.....))))))))))......((((....)))).....-....).)))))))..--......(((((.((((....))))....)--)))) ( -30.00) >DroPer_CAF1 107298 109 - 1 AAUUAUGGUAUCGCAUUGUUCAAAGUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAUAUUUCGGUGGGU-UUUCUCACACACUUUGAAGGGG---UAUUGCCCAUGAGAG ..(((((.(((.(((((((((.......))))))))).)))...((((.(((((............(((((.-...)))))............))---))))))))))))... ( -32.65) >consensus AAUUAUGGUAUCGCAUUGUUCAAAGCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU_UUUCGUUGGCA_AGG__CAUACGCUAGAAAAAGG___UAUGGAAAAUGAUAG ...((((.(((.(((((((((.......))))))))).)))..((((....)))).......................))))............................... (-18.58 = -18.92 + 0.33)

| Location | 17,584,154 – 17,584,256 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -16.11 |
| Energy contribution | -15.83 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
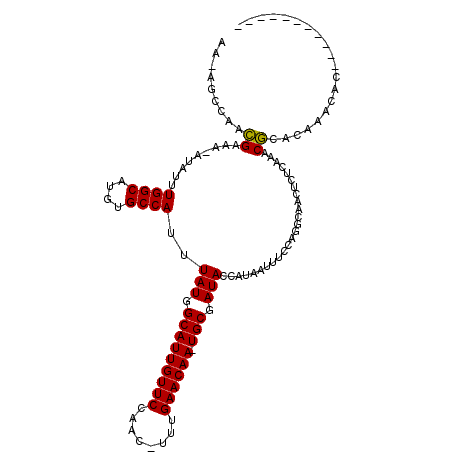
>2L_DroMel_CAF1 17584154 102 + 22407834 CUUUGCCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAGC-UUUGAACAA-UGCGAUACCAUAAUUUCCAGGCAACUCGCAAACGCACAAACUC----------- ..(((((...((((-(((..((((....))))..))).(((((((((....-...))))))-)))..........))))..)))))...((....))........----------- ( -28.10) >DroPse_CAF1 106897 113 + 1 AA-ACCCACCGAAAUAUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAAC-UUUGAACAA-UGCGAUACCAUAAUUUCCAGGCAGCUCUCUCUCGAACAAAUACAUAGGACACCA ..-..((.((....((((..((((....))))..))))(((((((((....-...))))))-)))................))..(.((......)).).........))...... ( -22.80) >DroEre_CAF1 99246 102 + 1 CUUUGCCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAGC-UUUGAACAA-UGCGAUACCAUAAUUUUCAGGCAACUCGCAAGCGCACAAACGC----------- ..(((((...((((-((...((((....))))..(((.(((((((((....-...))))))-))).))).....)))))).))))).......(((......)))----------- ( -29.90) >DroWil_CAF1 167665 103 + 1 -------AAUGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAACAUCUGAACAAUUGCGAUACCAUAAUUUUCAGGCA-CACACCAACACACAAUCAA----GCAUCCA -------..(((..-....((((..((((((.......(((((((((........)))))).)))((...........)).))))-))..)))).......))).----....... ( -22.62) >DroAna_CAF1 105659 102 + 1 GCUAGGCAACGAAA-AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAAG-UUGGAACAA-UGCGAUACCAUAGUUUCCAGGCAACUCUUGAGCGCACACACUC----------- (((.((.(((....-(((..((((....))))..))).((((((((((...-..)))))))-))).........))).)).))).......(((.(....).)))----------- ( -32.70) >DroPer_CAF1 107333 111 + 1 AA-ACCCACCGAAAUAUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAAC-UUUGAACAA-UGCGAUACCAUAAUUUCCAGGCAGCUCU--CUCGAACAAAUACAUAGGACACCA ..-.....((....((((..((((....))))..))))(((((((((....-...))))))-)))...........(((.(((......)--)).)))..........))...... ( -22.70) >consensus AA_AGCCAACGAAA_AUAUUUGGCAUGUGCCAUUUAUGGCAUUGUUCCAAC_UUUGAACAA_UGCGAUACCAUAAUUUCCAGGCAACUCUCAAACGCACAAACAC___________ .........((.........((((....))))..(((.(((((((((........)))))).))).))).........................)).................... (-16.11 = -15.83 + -0.28)
| Location | 17,584,154 – 17,584,256 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -19.06 |
| Energy contribution | -18.87 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
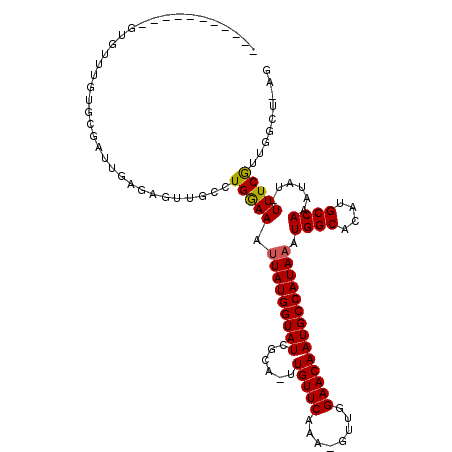
>2L_DroMel_CAF1 17584154 102 - 22407834 -----------GAGUUUGUGCGUUUGCGAGUUGCCUGGAAAUUAUGGUAUCGCA-UUGUUCAAA-GCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGGCAAAG -----------...(((((......)))))(((((..((((.(((..(((.(((-((((((...-....))))))))).)))..((((....)))).))).-))))...))))).. ( -29.30) >DroPse_CAF1 106897 113 - 1 UGGUGUCCUAUGUAUUUGUUCGAGAGAGAGCUGCCUGGAAAUUAUGGUAUCGCA-UUGUUCAAA-GUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAUAUUUCGGUGGGU-UU ......(((((......((((......))))......(((((.(((.(((.(((-((((((...-....))))))))).)))..((((....))))..)))))))).))))).-.. ( -28.60) >DroEre_CAF1 99246 102 - 1 -----------GCGUUUGUGCGCUUGCGAGUUGCCUGAAAAUUAUGGUAUCGCA-UUGUUCAAA-GCUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGGCAAAG -----------((((....)))).......((((((((((((.....(((.(((-((((((...-....))))))))).)))..((((....))))...))-)))))..))))).. ( -32.40) >DroWil_CAF1 167665 103 - 1 UGGAUGC----UUGAUUGUGUGUUGGUGUG-UGCCUGAAAAUUAUGGUAUCGCAAUUGUUCAGAUGUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCAUU------- .......----.(((..((((.((((((((-((((......(((((((((......(((((........))))))))))))))..))))))))))))))))-..)))..------- ( -34.90) >DroAna_CAF1 105659 102 - 1 -----------GAGUGUGUGCGCUCAAGAGUUGCCUGGAAACUAUGGUAUCGCA-UUGUUCCAA-CUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU-UUUCGUUGCCUAGC -----------(((((....)))))....(.((((.(....)...)))).)(((-(((((((..-...))))))))))......((((....)))).....-.............. ( -34.20) >DroPer_CAF1 107333 111 - 1 UGGUGUCCUAUGUAUUUGUUCGAG--AGAGCUGCCUGGAAAUUAUGGUAUCGCA-UUGUUCAAA-GUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAUAUUUCGGUGGGU-UU ......(((((......((((...--.))))......(((((.(((.(((.(((-((((((...-....))))))))).)))..((((....))))..)))))))).))))).-.. ( -30.40) >consensus ___________GUGUUUGUGCGAUUGAGAGUUGCCUGGAAAUUAUGGUAUCGCA_UUGUUCAAA_GUUGGAACAAUGCCAUAAAUGGCACAUGCCAAAUAU_UUUCGUUGGCU_AG ...................................(((((.(((((((((......(((((........)))))))))))))).((((....))))......)))))......... (-19.06 = -18.87 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:58 2006