
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,569,308 – 17,569,400 |
| Length | 92 |
| Max. P | 0.995943 |

| Location | 17,569,308 – 17,569,400 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
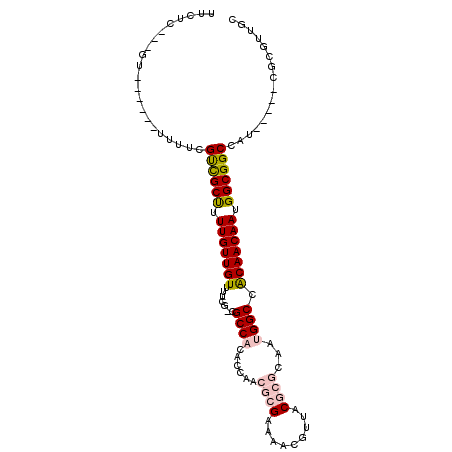
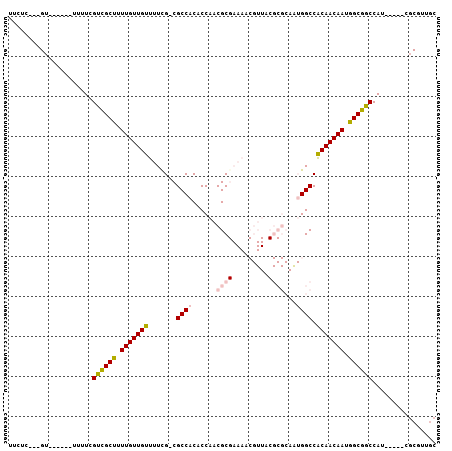
>2L_DroMel_CAF1 17569308 92 + 22407834 UUCUU---GU------UUUUCGUCGCUUUUGUUGUUUUCG-CGCCACACCAACGCGAAAACGUUACGCGCAAUGGCCACAACAAUGGCGGCCAU-----CGCGUUGC .....---..------.................(((((((-((.........)))))))))((.(((((..((((((.((....))..))))))-----))))).)) ( -29.50) >DroSec_CAF1 85654 92 + 1 UUCUU---GU------UUUUCGCCGCUUUUGUUGUUUUCG-CGCCACACCAACGCGAAAACGUUACGCGCAAUGGCCACAACAAUGGCGGCCAU-----CGCGUUGC .....---..------.................(((((((-((.........)))))))))((.(((((..((((((.((....))..))))))-----))))).)) ( -29.50) >DroSim_CAF1 84756 97 + 1 UUCUU---GU------UUUUCGUCGCUUUUGUUGUUUUCG-CGCCACACCAACGCGAAAACGUUACGCGCAAUGGCCACAACAAUGGCGGCCAUCGCAUCGCGUUGC .....---..------.................(((((((-((.........)))))))))((.((((((.((((((.((....))..)))))).))...)))).)) ( -29.50) >DroEre_CAF1 84365 74 + 1 UCCUC---GA------UUUUCGUCGCUUUUGUUGUUUUCG-CGCCACGCCAACGC------------------GGCCACAACAAUGGCGGCCAU-----CGCGUUGC ..(.(---((------(....((((((.(((((((..(((-((.........)))------------------))..))))))).)))))).))-----)).).... ( -25.30) >DroYak_CAF1 86101 74 + 1 UGCCC---GU------UUAUCGUCGCUUUUGUUGUUUUCG-CGCCACGCCAACGC------------------GGCCACAACAAUGGCGGCCAU-----CGCGUUGC .((.(---((------.....((((((.(((((((..(((-((.........)))------------------))..))))))).))))))...-----.)))..)) ( -25.00) >DroAna_CAF1 91328 98 + 1 CUCUCAGUGUUUUUAGUUUUUGCUGCCAUUGUUGUUUUUGCCGCCACGCCAACGCGAAACCGUUACGCGCAGUGGCCGCAACAAUGGCGGCCAU-----CGCC---- .....................((((((((((((((....(((((..(((.((((......))))..)))..))))).))))))))))))))...-----....---- ( -37.50) >consensus UUCUC___GU______UUUUCGUCGCUUUUGUUGUUUUCG_CGCCACACCAACGCGAAAACGUUACGCGCAAUGGCCACAACAAUGGCGGCCAU_____CGCGUUGC .....................((((((.(((((((.......((((......((((.........))))...)))).))))))).))))))................ (-18.69 = -19.47 + 0.78)

| Location | 17,569,308 – 17,569,400 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.97 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
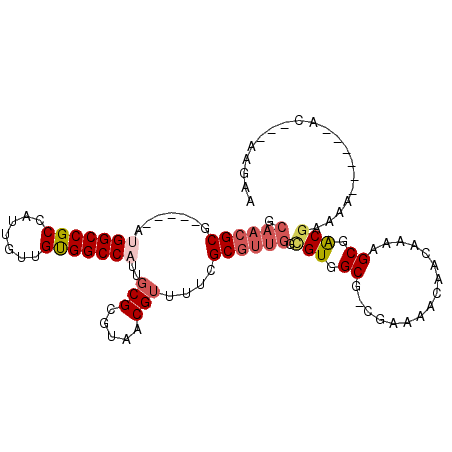
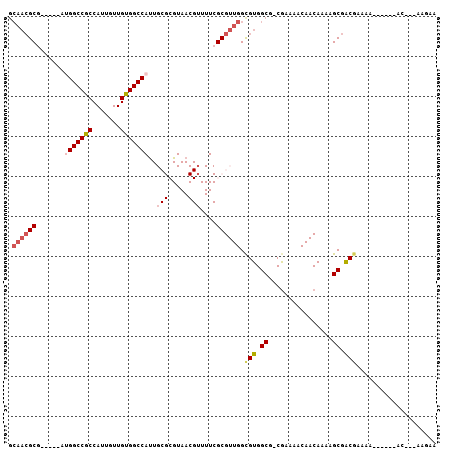
>2L_DroMel_CAF1 17569308 92 - 22407834 GCAACGCG-----AUGGCCGCCAUUGUUGUGGCCAUUGCGCGUAACGUUUUCGCGUUGGUGUGGCG-CGAAAACAACAAAAGCGACGAAAA------AC---AAGAA ((..((((-----((((((((.......))))))))))))......(((((((((((.....))))-))))))).......))........------..---..... ( -39.40) >DroSec_CAF1 85654 92 - 1 GCAACGCG-----AUGGCCGCCAUUGUUGUGGCCAUUGCGCGUAACGUUUUCGCGUUGGUGUGGCG-CGAAAACAACAAAAGCGGCGAAAA------AC---AAGAA ((..((((-----((((((((.......))))))))))))......(((((((((((.....))))-))))))).......))........------..---..... ( -39.40) >DroSim_CAF1 84756 97 - 1 GCAACGCGAUGCGAUGGCCGCCAUUGUUGUGGCCAUUGCGCGUAACGUUUUCGCGUUGGUGUGGCG-CGAAAACAACAAAAGCGACGAAAA------AC---AAGAA ((...(((.((((((((((((.......)))))))))))))))...(((((((((((.....))))-))))))).......))........------..---..... ( -41.60) >DroEre_CAF1 84365 74 - 1 GCAACGCG-----AUGGCCGCCAUUGUUGUGGCC------------------GCGUUGGCGUGGCG-CGAAAACAACAAAAGCGACGAAAA------UC---GAGGA ....(.((-----((.(.(((..(((((((.(((------------------(((....)))))).-.....)))))))..))).)....)------))---).).. ( -24.10) >DroYak_CAF1 86101 74 - 1 GCAACGCG-----AUGGCCGCCAUUGUUGUGGCC------------------GCGUUGGCGUGGCG-CGAAAACAACAAAAGCGACGAUAA------AC---GGGCA ((..((..-----((.(.(((..(((((((.(((------------------(((....)))))).-.....)))))))..))).).))..------.)---).)). ( -25.20) >DroAna_CAF1 91328 98 - 1 ----GGCG-----AUGGCCGCCAUUGUUGCGGCCACUGCGCGUAACGGUUUCGCGUUGGCGUGGCGGCAAAAACAACAAUGGCAGCAAAAACUAAAAACACUGAGAG ----(((.-----...)))((((((((((..(((.(..(((.(((((......))))))))..).))).....))))))))))........................ ( -40.00) >consensus GCAACGCG_____AUGGCCGCCAUUGUUGUGGCCAUUGCGCGUAACGUUUUCGCGUUGGCGUGGCG_CGAAAACAACAAAAGCGACGAAAA______AC___AAGAA .((((((.......(((((((.......)))))))..(((.....)))....)))))).(((.((................)).))).................... (-18.92 = -19.97 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:45 2006