| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,545,042 – 17,545,154 |
| Length | 112 |
| Max. P | 0.576207 |

| Location | 17,545,042 – 17,545,154 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
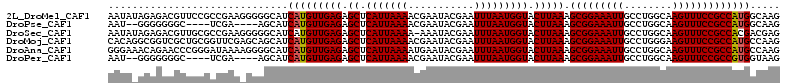
>2L_DroMel_CAF1 17545042 112 + 22407834 CUUGCCAUGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCGUUUUAAUGAGCUCUCAACAUGAUGCCCCCUUCGGCGGAACGUCUCUAUAUU .(((((.(((((....)..))))))))).(((((((..(((...(((((((..((....)).))))))).((((.......)).))...))).)))))))............ ( -30.40) >DroPse_CAF1 65143 102 + 1 CUUGCCAUGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCGUUUUAAUGAGCUCUCAACAUGAUGCU----UCGA----GCCCCCCC--AUU .(((((.(((((....)..))))))))).....((((.(((((.(((((((..((....)).))))))).....((.....))))))----).))----))......--... ( -26.10) >DroSec_CAF1 62034 111 + 1 CUCGUCGUGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUU-UUUUAAUGAGCUCUCAACAUGAUGCCCCCUUCGGCGCAACGUCUCUAUAUU ...(((((((((((((.((((...)))))))))))....(((..(((((((.........-.))))))).))).....))))))(((......)))................ ( -26.10) >DroMoj_CAF1 69204 112 + 1 CUUGGCAUGGCGGAAACUUCCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCGUUUUAAUGAGCUCUCAACAUGAUGCUGCUCGAACCGCAGCGACCGCCUGUG ....(((.(((((..((((...((((.......)))).))))..(((((((..((....)).)))))))..............((((((.......)))))).)))))))). ( -29.20) >DroAna_CAF1 70653 112 + 1 CUUGGCAUGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCAUUUUAAUGAGCUCUCAACAUGAUGCCCCUUUUAUCCCGGGUUCUGUUUCCC ...(((((((((((((.((((...))))))))))))...(((..(((((((...........))))))).))).........)))))...........(((........))) ( -25.90) >DroPer_CAF1 65411 102 + 1 CUUACCACGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCGUUUUAAUGAGCUCUCAACAUGAUGCU----UCGA----GCCCCCCC--AUU ..(((...((((((((.((((...))))))))))))....))).(((((((..((....)).))))))).((((.............----..))----))......--... ( -21.96) >consensus CUUGCCAUGGCGGAAACUUGCCAGGCAAUUUCCGCUUUAAGUACCAUUAAAUUCGUAUUCGUUUUAAUGAGCUCUCAACAUGAUGCCCC_UUCGACGCAGCGUCCCC_UAUU .....(((((((((((.((((...)))))))))))....(((..(((((((...........))))))).))).....)))).............................. (-17.82 = -18.02 + 0.19)


| Location | 17,545,042 – 17,545,154 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -21.74 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17545042 112 - 22407834 AAUAUAGAGACGUUCCGCCGAAGGGGGCAUCAUGUUGAGAGCUCAUUAAAACGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCAUGGCAAG ............(((((((....)..........(((((.((.(((((((.((....))..))))))))).))))).)))))).(((((((((........)))).))))). ( -31.30) >DroPse_CAF1 65143 102 - 1 AAU--GGGGGGGC----UCGA----AGCAUCAUGUUGAGAGCUCAUUAAAACGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCAUGGCAAG ..(--(((.((((----((..----(((.....)))..))))))((((((.((....))..))))))....)))).........(((((((((........)))).))))). ( -27.50) >DroSec_CAF1 62034 111 - 1 AAUAUAGAGACGUUGCGCCGAAGGGGGCAUCAUGUUGAGAGCUCAUUAAAA-AAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCACGACGAG ..........((((((.((....)).))......(((((.((.(((((((.-.........))))))))).))))).(((((((((((...)).)))))))))...)))).. ( -31.60) >DroMoj_CAF1 69204 112 - 1 CACAGGCGGUCGCUGCGGUUCGAGCAGCAUCAUGUUGAGAGCUCAUUAAAACGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGGAAGUUUCCGCCAUGCCAAG ....((((((.(((((.......))))))))((((((((.((.(((((((.((....))..))))))))).))))).(((((((((.(....).)))))))))))))))... ( -37.30) >DroAna_CAF1 70653 112 - 1 GGGAAACAGAACCCGGGAUAAAAGGGGCAUCAUGUUGAGAGCUCAUUAAAAUGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCAUGCCAAG (((........)))...........(((((....(((((.((.(((((((...........))))))))).))))).(((((((((((...)).))))))))).)))))... ( -30.60) >DroPer_CAF1 65411 102 - 1 AAU--GGGGGGGC----UCGA----AGCAUCAUGUUGAGAGCUCAUUAAAACGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCGUGGUAAG ...--....((((----((..----(((.....)))..)))))).(((..(((..(((.(......).)))......(((((((((((...)).))))))))))))..))). ( -28.20) >consensus AAUA_AGAGACGCUGCGCCGAA_GGAGCAUCAUGUUGAGAGCUCAUUAAAACGAAUACGAAUUUAAUGGUACUUAAAGCGGAAAUUGCCUGGCAAGUUUCCGCCAUGGCAAG ..............................(((((((((.((.(((((((...........))))))))).))))).(((((((((........)))))))))))))..... (-21.74 = -21.77 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:37 2006