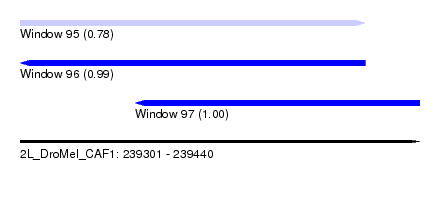
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 239,301 – 239,440 |
| Length | 139 |
| Max. P | 0.996186 |

| Location | 239,301 – 239,421 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
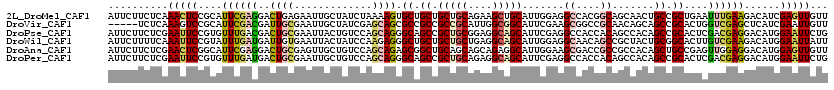
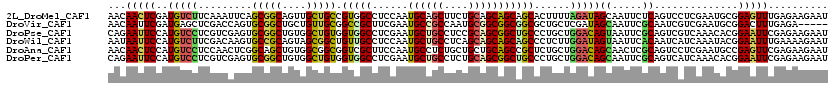
>2L_DroMel_CAF1 239301 120 + 22407834 AUUCUUCUCAAACUCCGCAUUCGAGGACUGAGAAUUGCUAUCUAAAAGUGCUGCUGCUGCAGAAGCUGCAUUGGAGGCCACGGCAGCAACUGCCGCUGAAUUUGAAGACAUCGAGUUGUU ....((((((...(((........))).))))))..(((.(((((.(((.((((....))))..)))...))))))))((((((((...)))))).))(((((((.....)))))))... ( -33.70) >DroVir_CAF1 36752 115 + 1 -----UCUCAAAGUCCGCAUUCGACGAUUGCGAAUUGCUAUCGAGCAGCGCCGCCGCCGCAUUGGCGGCAUUCGAAGCGGCCGCAACAGCAGCCGCACUGGUCGAGCUCAUCGAAUUGUU -----...........((((((((.(((.((.....)).)))((((..((((((((((.....)))))).......(((((.((....)).)))))...)).)).)))).))))).))). ( -45.70) >DroPse_CAF1 37980 120 + 1 AUUCUUCUCGAAUUCCGUGUUUGACGACUGCGAAUUACUGUCCAGCAGGGCAGCCGCUGCGGAGGCAGCAUUCGAGGCCACCACAGCCACAGCCGCACUCGACGAGGACAUGGAAUUCUG .........(((((((((((((..(((.((((.....((((((....))))))..((((.((.(((..(....)..))).)).))))......)))).)))....))))))))))))).. ( -50.20) >DroWil_CAF1 39756 120 + 1 AUUCUUUUCAAAUUCCGUAUUUGAUGAUUGUGAAUUACUAUCCAAGAGGGCUGCUGCUGCUGAGGCAGCAUUGGAGGCAACAGCCGCUACUGCGGCACUUGUCGAAGACAUGGAAUUAUU ..........((((((((.(((((..(.............(((((....((((((.(....).)))))).)))))(....).(((((....)))))..)..)))))...))))))))... ( -40.50) >DroAna_CAF1 37294 120 + 1 AUUCUUCUCGAACUCGGCAUUCGAGGACUGCGAGUUGCUGUCCAGCAGAGCGGCUGCAGCAGCAGAGGCAUUGGAAGCGACCGCCGCCACAGCUGCCGAGUUGGAGGACAUGGAGUUGUU (((((((((.(((((((((...(.((.(.(((.((((((.(((((....((..((((....))))..)).)))))))))))))).))).)...))))))))).))))....))))).... ( -51.00) >DroPer_CAF1 38397 120 + 1 AUUCUUCUCGAAUUCCGUGUUUGAUGACUGCGAAUUGCUGUCCAGCAGGGCAGCCGCUGCAGAGGCAGCAUUCGAGGCCACCACAGCCACAGCCGCACUCGACGAGGACAUGGAAUUCUG .........(((((((((((((..(((.(((((((.(((((((....))))))).(((((....)))))))))..(((.......)))......))).)))....))))))))))))).. ( -48.90) >consensus AUUCUUCUCAAAUUCCGCAUUCGACGACUGCGAAUUGCUAUCCAGCAGGGCAGCCGCUGCAGAGGCAGCAUUCGAGGCCACCGCAGCCACAGCCGCACUCGUCGAGGACAUGGAAUUGUU ..........(((((....((((((..((((.............)))).((.((.(((((....))))).......((....)).......)).))....))))))......)))))... (-17.03 = -16.95 + -0.08)
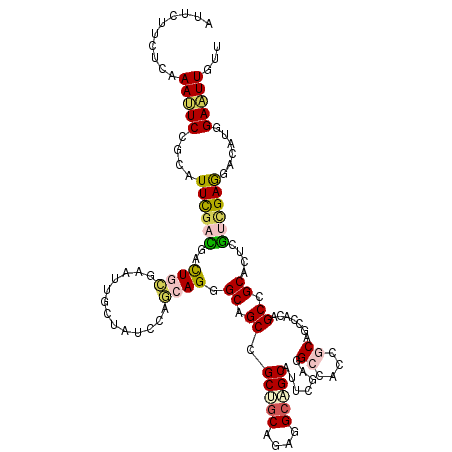
| Location | 239,301 – 239,421 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -24.13 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 239301 120 - 22407834 AACAACUCGAUGUCUUCAAAUUCAGCGGCAGUUGCUGCCGUGGCCUCCAAUGCAGCUUCUGCAGCAGCAGCACUUUUAGAUAGCAAUUCUCAGUCCUCGAAUGCGGAGUUUGAGAAGAAU .....(((((...((((..((((((.(((.((((((((..(((...))).(((((...)))))))))))))......(((.......)))..))))).))))..)))).)))))...... ( -34.80) >DroVir_CAF1 36752 115 - 1 AACAAUUCGAUGAGCUCGACCAGUGCGGCUGCUGUUGCGGCCGCUUCGAAUGCCGCCAAUGCGGCGGCGGCGCUGCUCGAUAGCAAUUCGCAAUCGUCGAAUGCGGACUUUGAGA----- ....(((((((((..((((.((((((((((((....))))))........(((((((.....))))))))))))).))))..((.....))..))))))))).............----- ( -52.70) >DroPse_CAF1 37980 120 - 1 CAGAAUUCCAUGUCCUCGUCGAGUGCGGCUGUGGCUGUGGUGGCCUCGAAUGCUGCCUCCGCAGCGGCUGCCCUGCUGGACAGUAAUUCGCAGUCGUCAAACACGGAAUUCGAGAAGAAU ..(((((((.(((....(.(((.((((((.((.(((((((.(((..........))).))))))).)).))).((((....))))....))).))).)..))).)))))))......... ( -49.80) >DroWil_CAF1 39756 120 - 1 AAUAAUUCCAUGUCUUCGACAAGUGCCGCAGUAGCGGCUGUUGCCUCCAAUGCUGCCUCAGCAGCAGCAGCCCUCUUGGAUAGUAAUUCACAAUCAUCAAAUACGGAAUUUGAAAAGAAU ............((((...(((((.((((((.((.(((((((((......(((((...)))))))))))))))).)))(((.((.....)).)))........))).)))))..)))).. ( -32.50) >DroAna_CAF1 37294 120 - 1 AACAACUCCAUGUCCUCCAACUCGGCAGCUGUGGCGGCGGUCGCUUCCAAUGCCUCUGCUGCUGCAGCCGCUCUGCUGGACAGCAACUCGCAGUCCUCGAAUGCCGAGUUCGAGAAGAAU ..............(((.(((((((((((((..(((((((..((.......))..)))))))..)))).........((((.((.....)).)))).....))))))))).)))...... ( -53.30) >DroPer_CAF1 38397 120 - 1 CAGAAUUCCAUGUCCUCGUCGAGUGCGGCUGUGGCUGUGGUGGCCUCGAAUGCUGCCUCUGCAGCGGCUGCCCUGCUGGACAGCAAUUCGCAGUCAUCAAACACGGAAUUCGAGAAGAAU ..(((((((........((((....)))).(((....(((((((..((((((((((....))))).((((.((....)).)))).)))))..)))))))..))))))))))......... ( -47.60) >consensus AACAAUUCCAUGUCCUCGACCAGUGCGGCUGUGGCUGCGGUGGCCUCCAAUGCUGCCUCUGCAGCAGCAGCCCUGCUGGACAGCAAUUCGCAGUCAUCAAAUACGGAAUUCGAGAAGAAU ...(((((..(((((.........(((((....))))).(((((......((((((....)))))))))))......)))))((.....))..............))))).......... (-24.13 = -23.42 + -0.72)
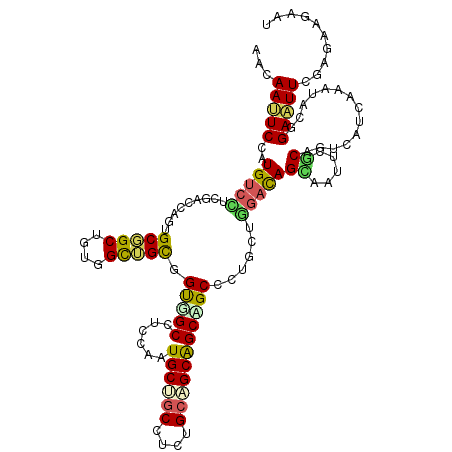
| Location | 239,341 – 239,440 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -22.44 |
| Energy contribution | -24.03 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
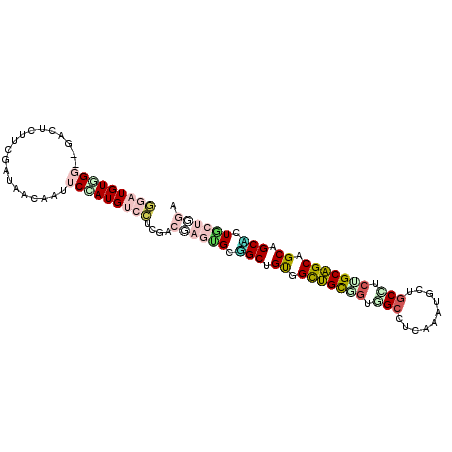
>2L_DroMel_CAF1 239341 99 - 22407834 AGAUGUUG---GUCUUUUUGAUAACAACUCGAUGUCUUCAAAUUCAGCGGCAGUUGCUGCCGUGGCCUCCAAUGCAGCUUCUGCAGCAGCAGCACUUUUAGA ...(((((---.....(((((..(((......)))..)))))....((.((((..(((((..(((...)))..)))))..)))).))..)))))........ ( -29.90) >DroPse_CAF1 38020 102 - 1 GGAUGUGGGGGGCCUGUUCGAUCAGAAUUCCAUGUCCUCGUCGAGUGCGGCUGUGGCUGUGGUGGCCUCGAAUGCUGCCUCCGCAGCGGCUGCCCUGCUGGA ((((((((((...(((......)))..))))))))))...((.((((.(((.((.(((((((.(((..........))).))))))).)).))).)))).)) ( -47.40) >DroGri_CAF1 37740 102 - 1 CGAUGUGGGCGGACUAUUCGAUAACAAUUCGAUGAGUUCGACGAGUGCUGCUGCAGUUGCAGCUGCCUCAAGUGCUGCAUCCGCAGCAGCAGCGUUGCUCGA .....((((((((((..((((.......))))..)))))((((..(((((((((.(.((((((..........)))))).).))))))))).))))))))). ( -47.80) >DroMoj_CAF1 35312 99 - 1 GGAUGUUGG---UCUCUUCGACAACAAUUCAAUGAGCUCGACAAGCGCUGCUGCCGUUGCAGCUGCCUCAAAUGCGGCCAAUGCAGCGGCAGCACUGCUCGA ...(((((.---.(((...((.......))...)))..)))))((((.((((((((((((((((((.......)))))...))))))))))))).))))... ( -44.10) >DroAna_CAF1 37334 99 - 1 GGAUGUUG---GACUCUUCGACAACAACUCCAUGUCCUCCAACUCGGCAGCUGUGGCGGCGGUCGCUUCCAAUGCCUCUGCUGCUGCAGCCGCUCUGCUGGA ((((((((---..........)))))..)))......((((.(..(((.((((..(((((((..((.......))..)))))))..)))).)))..).)))) ( -42.40) >DroPer_CAF1 38437 102 - 1 GGAUGUGGGGGGCCUGUUCGAUCAGAAUUCCAUGUCCUCGUCGAGUGCGGCUGUGGCUGUGGUGGCCUCGAAUGCUGCCUCUGCAGCGGCUGCCCUGCUGGA ((((((((((...(((......)))..))))))))))...((.((((.(((.((.((((..(.(((..........))).)..)))).)).))).)))).)) ( -44.60) >consensus GGAUGUGGG__GACUCUUCGAUAACAAUUCCAUGUCCUCGACGAGUGCGGCUGUGGCUGCGGUGGCCUCAAAUGCUGCCUCUGCAGCAGCAGCACUGCUGGA (((((((((...................)))))))))....((((((.(((.((.(((((((.(((..........))).))))))).)).))).)))))). (-22.44 = -24.03 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:48 2006