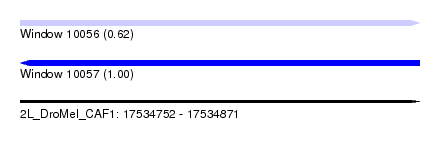
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,534,752 – 17,534,871 |
| Length | 119 |
| Max. P | 0.999568 |

| Location | 17,534,752 – 17,534,871 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.52 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.623968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17534752 119 + 22407834 GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUGGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCAGUUCCCACCUAUUGAACCCAUUUGUAGCCGCUCGC ((((((((((((((.(((......(((((...)))))..(((((((((....((.......))...))))))))).............))).)))))....)))))))..))....... ( -22.80) >DroSec_CAF1 51877 119 + 1 GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCGUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUAUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCCGCUCGC ((((((((((((((.(((...(((((((..........((((((((((....((.......))...))))))))))....))))))).))).)))))....)))))))..))....... ( -25.94) >DroSim_CAF1 51018 119 + 1 GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCCGCUCGC ((((((((((((((.(((...............((((.(((((...))))).))))...(((((((..((.....))..)))))))..))).)))))....)))))))..))....... ( -27.30) >DroEre_CAF1 53043 109 + 1 GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUCAAACCA----------CUCGU ........((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).))))))......----------..... ( -23.60) >DroYak_CAF1 52925 119 + 1 GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUGAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGCAGCCCCUCGU ((((((((((((((.(((...............(((..(((((...)))))..)))...((((((((((....)))...)))))))..))).)))))....)))))))..))....... ( -25.60) >consensus GCCAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCCGCUCGC ..((((((((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).)))))....)))))))........... (-22.48 = -23.52 + 1.04)


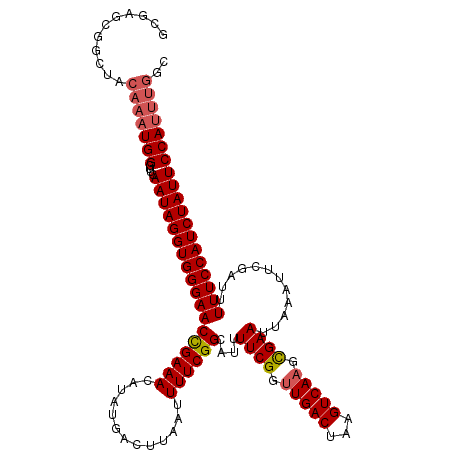
| Location | 17,534,752 – 17,534,871 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -30.60 |
| Energy contribution | -31.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17534752 119 - 22407834 GCGAGCGGCUACAAAUGGGUUCAAUAGGUGGGAACUGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACCAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC ((.....))..((((((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............))))))))))))))))))).. ( -33.00) >DroSec_CAF1 51877 119 - 1 GCGAGCGGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAAUAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCACGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC ((.....))..((((((((....(((((((((((..........(((((((....(((((.......))))).)))))))..(((((...)))))...))))))))))))))))))).. ( -32.60) >DroSim_CAF1 51018 119 - 1 GCGAGCGGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC ((.....))..((((((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............))))))))))))))))))).. ( -35.20) >DroEre_CAF1 53043 109 - 1 ACGAG----------UGGUUUGAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC .((((----------(((....((((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............))))))))))))))))))).. ( -32.60) >DroYak_CAF1 52925 119 - 1 ACGAGGGGCUGCAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUCAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC ...........((((((((....(((((((((((((((((...(((....)))))))))....((((.(((((...))))).))))............))))))))))))))))))).. ( -34.50) >consensus GCGAGCGGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUGGC ...........(((((((....((((((((((((((((((.............))))))....((((.(((((...))))).))))............))))))))))))))))))).. (-30.60 = -31.20 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:31 2006