
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,529,630 – 17,529,771 |
| Length | 141 |
| Max. P | 0.964641 |

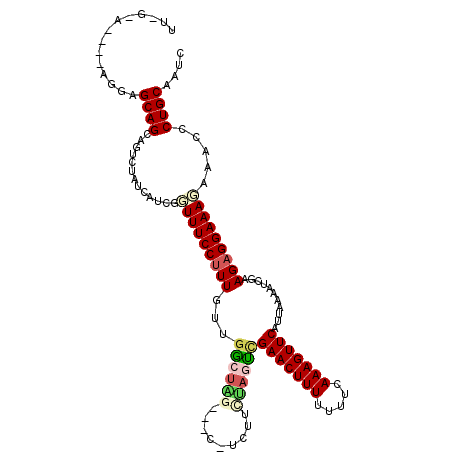
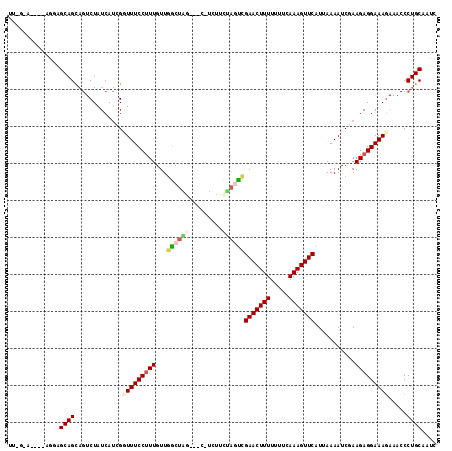
| Location | 17,529,630 – 17,529,738 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -17.21 |
| Energy contribution | -18.21 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17529630 108 + 22407834 UUUGGA----AGGAGCAGCAGUCUAUCAUCGGUUUCCUUUGUUGGCUAA----CUCUUCUAGCCGAACUUUUUUCCAAAGUUCAUUAAAAUCGAAGAGGAAAGAAACCCUGCAAUC ......----....((((...(((.((.(((((((........(((((.----......)))))(((((((.....)))))))....)))))))....)).)))....)))).... ( -26.60) >DroSec_CAF1 47213 107 + 1 -----A----AGAAGCAGCAGUCUAUCAUCGCUUUCCUUUGUUGGCUGGAAGCGUUUUUUAGUCGAACUUUUUUUCAAAGUUCAUUAAAAUCGAAGAGGAAAGAAACCCUGCAAUC -----.----.......((((..........(((((((((...(((((((((...)))))))))(((((((.....)))))))...........))))))))).....)))).... ( -26.46) >DroSim_CAF1 46024 107 + 1 -----A----AGGAGCAGCAGUCUAUCAUCGCUUUCCUUUGUUGGCUAGAAGCGUUUUCUAGUCGAACUUUUUUUAAAAGUUCAUUAAAAUCGAAGAGGAAAGAAACCCUGCAAUC -----.----.......((((..........(((((((((...(((((((((...)))))))))(((((((.....)))))))...........))))))))).....)))).... ( -29.66) >DroEre_CAF1 48006 104 + 1 UUAGGAAAUGGA-GGCAGU---CUAUCAACGGUUUCCUUUGUUGACUUG---C-GCCUCUACUUGAACUUUUUUUCAAAGUUCAUUAAAAUCUAAGAGGAAAG----CCUGCAAUC (((((...((((-(((.((---...(((((((......)))))))...)---)-)))))))..((((((((.....))))))))......))))).(((....----)))...... ( -27.60) >DroYak_CAF1 47811 108 + 1 UUUGAAAAAGAGGAGCAGCAG-CUAUCAUGGGUUUCCCUUGUUCGCUUA---C-GCUUCAUAUCGAACUUUUUCUUAAAGUUCAUUAAAAUCUAAGAGGAAAC---CCCUGCAAUC .................((((-.......((((((((((((...((...---.-))........(((((((.....))))))).........)))).))))))---)))))).... ( -27.61) >consensus UU_G_A____AGGAGCAGCAGUCUAUCAUCGGUUUCCUUUGUUGGCUAG___C_UCUUCUAGUCGAACUUUUUUUCAAAGUUCAUUAAAAUCGAAGAGGAAAGAAACCCUGCAAUC ..............((((.............(((((((((...((((((.........))))))(((((((.....)))))))...........))))))))).....)))).... (-17.21 = -18.21 + 1.00)

| Location | 17,529,630 – 17,529,738 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.65 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17529630 108 - 22407834 GAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGGAAAAAAGUUCGGCUAGAAGAG----UUAGCCAACAAAGGAAACCGAUGAUAGACUGCUGCUCCU----UCCAAA ....(((((((((((((................(((((((.....)))))))((((((.....----))))))....)))))))))(........)..))))....----...... ( -27.30) >DroSec_CAF1 47213 107 - 1 GAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGAAAAAAAGUUCGACUAAAAAACGCUUCCAGCCAACAAAGGAAAGCGAUGAUAGACUGCUGCUUCU----U----- ((..((((((((((((((((......(((((.((((((((.....))))))))..)))))...((.....))......))))))).......))))).)))).)).----.----- ( -23.11) >DroSim_CAF1 46024 107 - 1 GAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUUAAAAAAAGUUCGACUAGAAAACGCUUCUAGCCAACAAAGGAAAGCGAUGAUAGACUGCUGCUCCU----U----- ....((((((((((((((((.............(((((((.....)))))))(.((((((.....)))))).).....))))))).......))))).))))....----.----- ( -25.71) >DroEre_CAF1 48006 104 - 1 GAUUGCAGG----CUUUCCUCUUAGAUUUUAAUGAACUUUGAAAAAAAGUUCAAGUAGAGGC-G---CAAGUCAACAAAGGAAACCGUUGAUAG---ACUGCC-UCCAUUUCCUAA ((.((.(((----(((((((............((((((((.....))))))))......(((-.---...))).....))))))..(((....)---)).)))-).))..)).... ( -25.50) >DroYak_CAF1 47811 108 - 1 GAUUGCAGGG---GUUUCCUCUUAGAUUUUAAUGAACUUUAAGAAAAAGUUCGAUAUGAAGC-G---UAAGCGAACAAGGGAAACCCAUGAUAG-CUGCUGCUCCUCUUUUUCAAA ((..((((((---((((((.(((.........((((((((.....)))))))).......((-.---...))....)))))))))))..(....-)..))))...))......... ( -29.30) >consensus GAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGAAAAAAAGUUCGACUAGAAGA_G___CAAGCCAACAAAGGAAACCGAUGAUAGACUGCUGCUCCU____U_C_AA ....((((((((((((((((............((((((((.....)))))))).................(....)..))))))).......))))).)))).............. (-14.58 = -15.65 + 1.07)

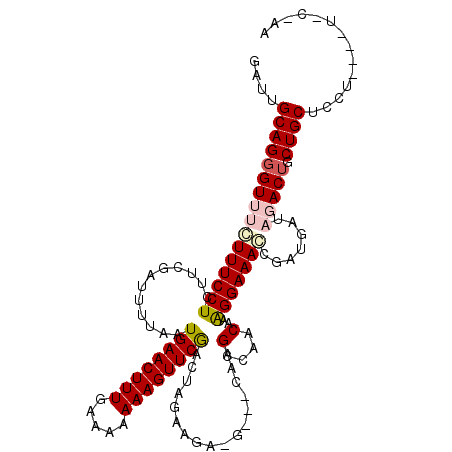
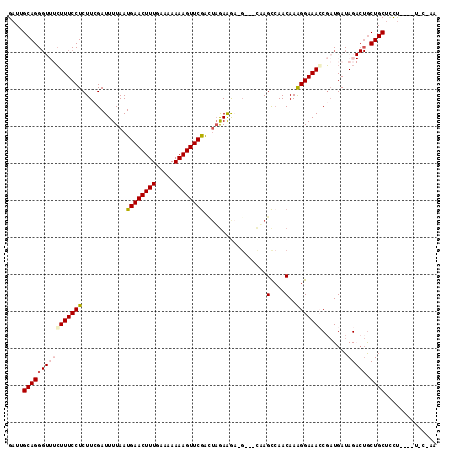
| Location | 17,529,666 – 17,529,771 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.89 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17529666 105 - 22407834 AAGCAAGCGUAGAACUAAUUUUAGUUAGCCAGAGAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGGAAAAAAGUUCGGCUAGAAGAG----UUAGCCAAC ..............((((((((..((((((.(((((((.((((........)))).)))))))...(((((((.....))))))))))))).))))----))))..... ( -28.50) >DroSec_CAF1 47244 109 - 1 AAGCAAGCGUAGAACUAAUUUUAGUUAGCCAGAGAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGAAAAAAAGUUCGACUAAAAAACGCUUCCAGCCAAC ..(.((((((........((((((((.....(((((((.((((........)))).)))))))...(((((((.....))))))))))))))).)))))).)....... ( -24.90) >DroSim_CAF1 46055 109 - 1 AAGCAAGCGUAGAACUAAUUUUAGUUAACCAGAGAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUUAAAAAAAGUUCGACUAGAAAACGCUUCUAGCCAAC .((.((((((........((((((((.....(((((((.((((........)))).)))))))...(((((((.....))))))))))))))).))))))))....... ( -24.30) >DroEre_CAF1 48042 101 - 1 AAGCUCGCGUAGAACUAAUUUUAGUUAGCCAGAGAUUGCAGG----CUUUCCUCUUAGAUUUUAAUGAACUUUGAAAAAAAGUUCAAGUAGAGGC-G---CAAGUCAAC ..(((.((....(((((....))))).(((.((((((..(((----....)))....))))))..((((((((.....))))))))......)))-)---).))).... ( -22.90) >DroYak_CAF1 47850 102 - 1 AAGCAAGCGUAGAACUUAUUUUAGUUAGCCAGAGAUUGCAGGG---GUUUCCUCUUAGAUUUUAAUGAACUUUAAGAAAAAGUUCGAUAUGAAGC-G---UAAGCGAAC ..((..((((..((((......)))).....((((((..((((---(...)))))..))))))..((((((((.....)))))))).......))-)---)..)).... ( -20.40) >consensus AAGCAAGCGUAGAACUAAUUUUAGUUAGCCAGAGAUUGCAGGGUUUCUUUCCUCUUCGAUUUUAAUGAACUUUGAAAAAAAGUUCGACUAGAAGA_G___CAAGCCAAC ......((..........((((((((.....(((((((.((((........)))).)))))))...(((((((.....)))))))))))))))..........)).... (-17.61 = -17.89 + 0.28)

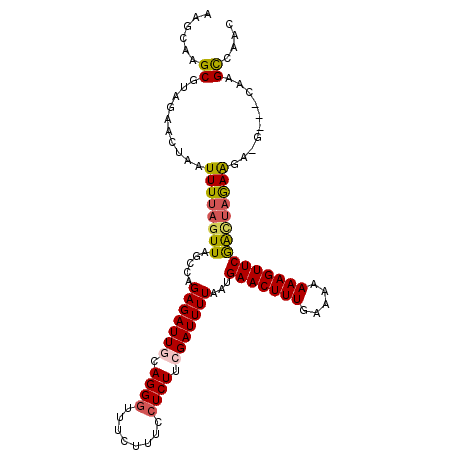
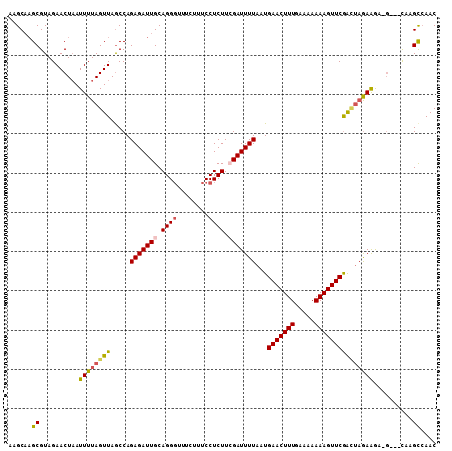
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:27 2006