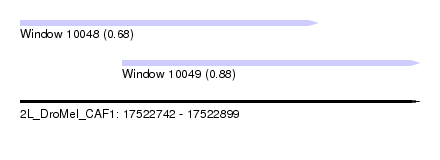
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,522,742 – 17,522,899 |
| Length | 157 |
| Max. P | 0.878682 |

| Location | 17,522,742 – 17,522,859 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -23.23 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
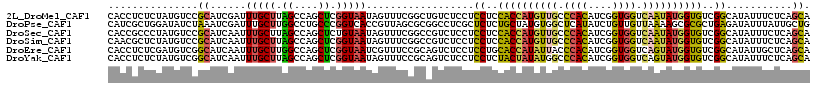
>2L_DroMel_CAF1 17522742 117 + 22407834 CACCUCUCUAUGUCCGCAUCGAUUUGCUUAGCCAGCUCGGUAAUAGUUUCGGCUGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCA ........((((((.(((......)))..((.((((.(((........))))))).)).......((((((((((.((((....)))).))))))))))..)))))).......... ( -35.60) >DroPse_CAF1 41774 117 + 1 CAUCGCUGGAUAUCUAAAUCGAUUUGCUUGGCCUGCCCGGUCACCGUUAGCGCGGCCUCGCUCUCUGCUAUGUGGCUCAUAUCUGUUGUUAAAAGCGCGCUGAGAUAUUUAUUGCUG ....(((((((((((.............(((((.....)))))....((((((((((.(((.....))...).)))).......(((......))))))))))))))))))..)).. ( -30.40) >DroSec_CAF1 39201 117 + 1 CACCGCCCUAUGUCCGCAUCAAUUUGCUUAGCCAGCUCUGUAAUAGUUUCGGCCGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCA ....(((..(((.(((.......((((..((....))..))))......))).))).........((((((((((.((((....)))).))))))))))..)))............. ( -32.32) >DroSim_CAF1 39102 117 + 1 CAACGCUCUAUGUCCGCAUCAAUUUGCUUAGCCAGCUCGGUAAUAGUUUCGGCCGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCA ....(((........(((......)))...(((.((.(((........)))))............((((((((((.((((....)))).))))))))))..))).........))). ( -34.90) >DroEre_CAF1 40370 117 + 1 CACCUCUCGAUGUCGGCAUCAAUUUGCUUGGCCAGCUCGGUAAUCGUUUCCGCAGUCUCCUCCUGCACCAUAUUACCCACAUCGGUGGUCAGUAUGGUGUCGGCAUAUUGCUCAGCA ........((((....))))....((((.(((..((((((.........))).))).....((.((((((((((..((((....))))..)))))))))).))......))).)))) ( -37.20) >DroYak_CAF1 39967 117 + 1 CACCUCUCUAUGUCGGCAUCAAUUUGCUUAGCCAGCUCGGUAAUAGUUUCCGCAGUCUCCUCCUCUACUAUAUGGCCCACAUCGGUGGUCAGUAUGGUGUCGGCAUAUUUCUCAGCA ........(((((((((((((...((((..((((((((((.........))).)))................))))((((....))))..))))))))))))))))).......... ( -30.59) >consensus CACCGCUCUAUGUCCGCAUCAAUUUGCUUAGCCAGCUCGGUAAUAGUUUCCGCAGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCA ...............((......(((((.((....)).)))))..................((..((((((((((.((((....)))).))))))))))..))...........)). (-23.23 = -22.60 + -0.63)
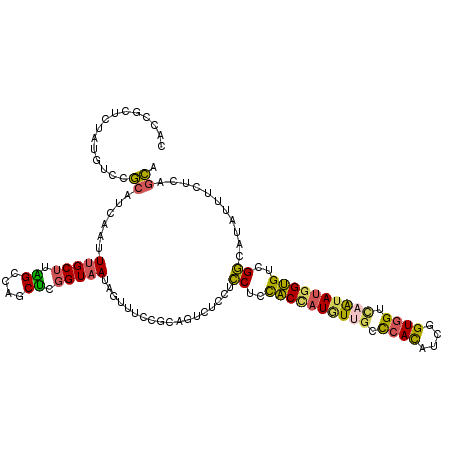

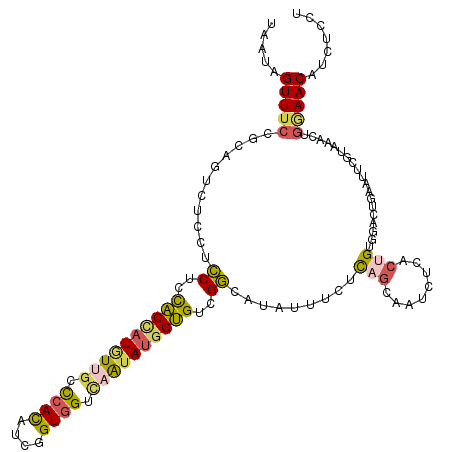
| Location | 17,522,782 – 17,522,899 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -20.16 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17522782 117 + 22407834 UAAUAGUUUCGGCUGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCAAUCUCACUGUGCACUGAAUUUGUAAACUGAAACAUCUCCU .....(((((((.........((..((((((((((.((((....)))).))))))))))..))...........((((..(((........)))..))))...)))))))....... ( -34.50) >DroPse_CAF1 41814 117 + 1 UCACCGUUAGCGCGGCCUCGCUCUCUGCUAUGUGGCUCAUAUCUGUUGUUAAAAGCGCGCUGAGAUAUUUAUUGCUGACCUGAUAGUGGGCCCCAAUACUCUUCUGGAACAAUUCUU ...((...(((((((((.(((.....))...).)))).......(((......))))))))(((.((((....(((.((......)).)))...)))))))....)).......... ( -25.20) >DroSec_CAF1 39241 117 + 1 UAAUAGUUUCGGCCGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCAAUCUCACUGUGUACUGAAUUCGUAAACUGGAACAUCUCCU ....(((((((((((..........((((((((((.((((....)))).)))))))))).)))).......((((....(.....)....))))...)).)))))(((.....))). ( -33.00) >DroSim_CAF1 39142 117 + 1 UAAUAGUUUCGGCCGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCAAUCUCACUGUGUACUGAAUUCGUAAACUGGAACAUCUCCU ....(((((((((((..........((((((((((.((((....)))).)))))))))).)))).......((((....(.....)....))))...)).)))))(((.....))). ( -33.00) >DroEre_CAF1 40410 117 + 1 UAAUCGUUUCCGCAGUCUCCUCCUGCACCAUAUUACCCACAUCGGUGGUCAGUAUGGUGUCGGCAUAUUGCUCAGCAAUCUCACCGUGGAUUGCAUUCGUAUGCUGGAACAUCUCCU .....(((((.(((.......((.((((((((((..((((....))))..)))))))))).))...........(((((((......))))))).......))).)))))....... ( -39.30) >DroYak_CAF1 40007 117 + 1 UAAUAGUUUCCGCAGUCUCCUCCUCUACUAUAUGGCCCACAUCGGUGGUCAGUAUGGUGUCGGCAUAUUUCUCAGCAAUCUCACUGUGGAUGGCAUUCGUAUGCUGGAACAUCUCCU ........((((((((.........((((((((.(.((((....)))).).))))))))...((..........))......)))))))).(((((....)))))(((.....))). ( -30.00) >consensus UAAUAGUUUCCGCAGUCUCCUCCUCCACCAUGUUGCCCACAUCGGUGGUCAAUAUGGUGUCGGCAUAUUUCUCAGCAAUCUCACUGUGGACUGAAUUCGUAAACUGGAACAUCUCCU .....(((((...........((..((((((((((.((((....)))).))))))))))..)).........(((........)))...................)))))....... (-20.16 = -20.25 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:23 2006