
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
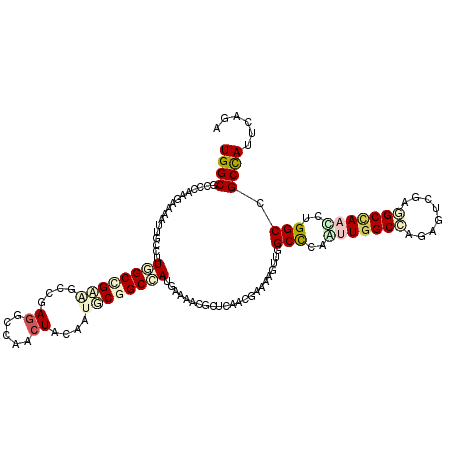
| Location | 17,508,852 – 17,508,972 |
| Length | 120 |
| Max. P | 0.828488 |

| Location | 17,508,852 – 17,508,972 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -24.26 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17508852 120 + 22407834 UGGCGCCCAAGAAAUUGGCUUUGGCCGAGGCCGAGGCCACCUAUAAUGCGGCCAUGAAGACGCUCAACGAAAAACUGGCCAUGUUGGCCAAAGUGGAGGCCAACCUGGCCGCCAUCCAGA (((((((((......((((((((((....)))))))))).......)).)))........................(((((.(((((((........))))))).)))))))))...... ( -53.02) >DroVir_CAF1 29442 120 + 1 UGGCGCCCAAGAAAAUUGCUUUGGCUGCAGCCGAGGCCGACUACAAUGCGGCCAUGAAACUGCUCAACGAGAAGUUGGCCCAACUGGCCCGCGUCGAAGCGAAUCUGGCGGCCAUUCAAA (((((((..(((...((((((((((.((((.((.(((((.(......)))))).))...))))..........((.((((.....)))).)))))))))))).)))))).))))...... ( -47.50) >DroGri_CAF1 34337 120 + 1 UGGCACCCAAGAAGAUUGCCCUCGCUGCCGCAGAGGCAGACUACAAUGCGGCCAUGAAACUGCUCAACGAGAAGUUGGCCCAACUGGCUCGAGUCGAAGCCAAUCUGGCCGCCAUUCAGA .((((..(.....)..))))....(((((.....)))))........(((((((.((....((.((((.....)))))).....(((((((...)).))))).)))))))))........ ( -40.30) >DroWil_CAF1 72053 120 + 1 UGGCACCCAAGAAAAUUGCAUUGGCCGAGGCGGAGGCCACAUAUAAUUCUGCAAUGAAAACGCUAAAUGAAAAGUUGGCUCAAUUGGCACGAGUUGAGGCUAACUUGGCUGCUAUACAAA (((((.(((((.........((((((..(((....))).....(((((((((..(((....(((((.(....).))))))))....))).)))))).))))))))))).)))))...... ( -34.80) >DroMoj_CAF1 30731 120 + 1 UGGCGCCCAAGAAGAUCGCCUUAGCUGCCGCCGAGGCCGACUACAAUGCGGCUAUGAAGUUGCUCAACGAAAAGCUAGCUCAAUUGGCCAGGGUCGAGGCCAACUUGGCCGCUAUUCAGA (((((.(((((..(...((((..(((...((((((((((.(......)))))).(((.((((((........))).)))))).)))))...)))..)))))..))))).)))))...... ( -42.70) >DroAna_CAF1 26934 120 + 1 UGGCGCCCAAGAAGAUUGCCCUGGCCGAAGCCGAGGCUACCUACAAUGCCGCCAUGAAGACGCUCAACGAAAAGUUGGCCCAGUUGGCCAGGGUGGAGGCCAAUCUGGCAGCCAUCCAGA ((((...........(..((((((((((.((((((((..........)))....(((......)))........)))))....))))))))))..)..(((.....))).))))...... ( -45.50) >consensus UGGCGCCCAAGAAAAUUGCCUUGGCCGAAGCCGAGGCCAACUACAAUGCGGCCAUGAAAACGCUCAACGAAAAGUUGGCCCAAUUGGCCAGAGUCGAGGCCAACCUGGCCGCCAUUCAGA ((((.................((((((((....((.....))....)))))))).......................(((..(((((((........)))))))..))).))))...... (-24.26 = -25.48 + 1.23)


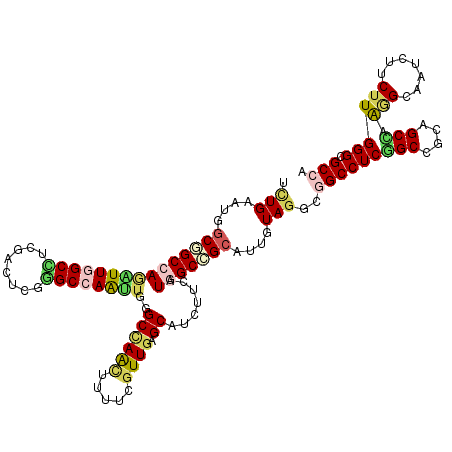
| Location | 17,508,852 – 17,508,972 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -45.63 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17508852 120 - 22407834 UCUGGAUGGCGGCCAGGUUGGCCUCCACUUUGGCCAACAUGGCCAGUUUUUCGUUGAGCGUCUUCAUGGCCGCAUUAUAGGUGGCCUCGGCCUCGGCCAAAGCCAAUUUCUUGGGCGCCA ......(((((((((.(((((((........))))))).)))))....(((.((((((.(((.....((((((.......))))))..))))))))).)))(((.........))))))) ( -53.80) >DroVir_CAF1 29442 120 - 1 UUUGAAUGGCCGCCAGAUUCGCUUCGACGCGGGCCAGUUGGGCCAACUUCUCGUUGAGCAGUUUCAUGGCCGCAUUGUAGUCGGCCUCGGCUGCAGCCAAAGCAAUUUUCUUGGGCGCCA .......(((((..(((((.(((.(((((..((((.....)))).......)))))))))))))..)))))((..((((((((....))))))))(((.(((.......))).))))).. ( -43.40) >DroGri_CAF1 34337 120 - 1 UCUGAAUGGCGGCCAGAUUGGCUUCGACUCGAGCCAGUUGGGCCAACUUCUCGUUGAGCAGUUUCAUGGCCGCAUUGUAGUCUGCCUCUGCGGCAGCGAGGGCAAUCUUCUUGGGUGCCA ..........((((.(((((((((......))))))))).)))).....(((((((.((((...((.(((..(...)..)))))...))))..)))))))((((..(.....)..)))). ( -48.10) >DroWil_CAF1 72053 120 - 1 UUUGUAUAGCAGCCAAGUUAGCCUCAACUCGUGCCAAUUGAGCCAACUUUUCAUUUAGCGUUUUCAUUGCAGAAUUAUAUGUGGCCUCCGCCUCGGCCAAUGCAAUUUUCUUGGGUGCCA ........(((.(((((...............((.((.((((.......)))).)).))......((((((..........(((((........))))).))))))...))))).))).. ( -27.70) >DroMoj_CAF1 30731 120 - 1 UCUGAAUAGCGGCCAAGUUGGCCUCGACCCUGGCCAAUUGAGCUAGCUUUUCGUUGAGCAACUUCAUAGCCGCAUUGUAGUCGGCCUCGGCGGCAGCUAAGGCGAUCUUCUUGGGCGCCA ......((((.(((.((((((((........))))))))..(((.((((......)))).........(((((......).))))...)))))).)))).((((.(((....))))))). ( -44.70) >DroAna_CAF1 26934 120 - 1 UCUGGAUGGCUGCCAGAUUGGCCUCCACCCUGGCCAACUGGGCCAACUUUUCGUUGAGCGUCUUCAUGGCGGCAUUGUAGGUAGCCUCGGCUUCGGCCAGGGCAAUCUUCUUGGGCGCCA ..((((.(((..((((.((((((........))))))))))((((((.....)))).)))))))))(((((.(...(.((((.(((..(((....)))..))).)))).)...).))))) ( -56.10) >consensus UCUGAAUGGCGGCCAGAUUGGCCUCGACUCGGGCCAAUUGGGCCAACUUUUCGUUGAGCAUCUUCAUGGCCGCAUUGUAGGCGGCCUCGGCCGCAGCCAAGGCAAUCUUCUUGGGCGCCA .(((....(((((((((((((((........))))))))..((((((.....)))).)).......)))))))....)))..(((((((((....))).(((.......)))))).))). (-29.14 = -29.62 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:18 2006