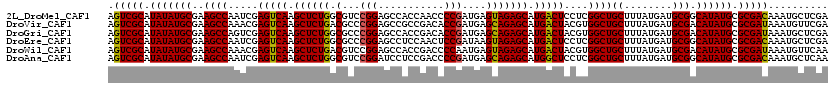
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,507,029 – 17,507,149 |
| Length | 120 |
| Max. P | 0.982452 |

| Location | 17,507,029 – 17,507,149 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -48.57 |
| Consensus MFE | -40.77 |
| Energy contribution | -40.72 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
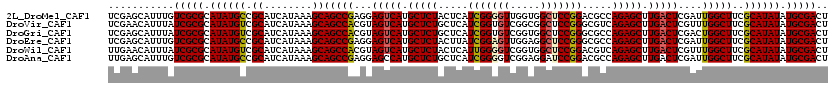
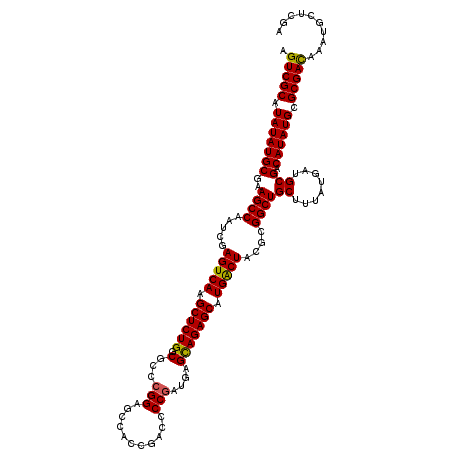
>2L_DroMel_CAF1 17507029 120 + 22407834 UCGAGCAUUUGUCGCGCAUAUGCCGCAUCAUAAAGCAGCCGAGGAGUCAUGCUCUACUCAUCGGGGUUGGUGGCUCCGGACGCCAGAGCUUGACUCGAUUGGCUUCGCAUAUAUGCGACU ..........((((((.((((((.((........))((((((.((((((.(((((.....(((((((.....))))))).....))))).))))))..))))))..)))))).)))))). ( -52.40) >DroVir_CAF1 27739 120 + 1 UCGAACAUUUAUCGCGCAUAUGUCGCAUCAUAAAGCAGCCACGUAGUCAUGCUCUGCUCAUCGGUGUCGGCGGCUCCGGGCGUCAGAGCUUGACUCGUUUGGCUUCGCAUAUAUGCGACU ...........(((((.((((((.((........))(((((((.(((((.((((((....((((.((.....)).))))....)))))).))))))))..))))..)))))).))))).. ( -45.50) >DroGri_CAF1 32059 120 + 1 UCGAGCAUUUAUCGCGCAUAUGUCGCAUCAUAAAGCAGCCACGUAGUCAUGCUCUGCUCAUCGGUGUCGGUGGCUCCGGGCGCCAGAGCUUGACUCGACUGGCUUCGCAUAUAUGCGACU .(((.......)))((((((((((((........))(((((((.(((((.((((((....((((.((.....)).))))....)))))).)))))))..)))))..).)))))))))... ( -45.40) >DroEre_CAF1 24413 120 + 1 UCGAGCAUUUGUCGCGCAUAUGCCGCAUCAUAAAGCAGCCGAGGAGUCAUGCUCUACUUAUCGGAGUUGGAGGCUCCGGGCGCCAGAGCUUGACUCGAUUGGCUUCGCAUAUAUGCGACU ..........((((((.((((((.((........))((((((.((((((.(((((.....(((((((.....))))))).....))))).))))))..))))))..)))))).)))))). ( -54.40) >DroWil_CAF1 70348 120 + 1 UUGAACAUUUAUCGCGCAUAUGUCGCAUCAUAAAGCAGCCACGUAGUCAUGCUCUACUCAUUGGGGUCGGUGGCUCCGGACGUCAGAGCUUGACUCGUUUGGCUUCGCAUAUAUGCGACU ...........(((((.((((((.((........))(((((((.(((((.(((((((((...(((((.....))))).)).)).))))).))))))))..))))..)))))).))))).. ( -43.50) >DroAna_CAF1 25151 120 + 1 UUGAGCAUUUGUCGCGCAUAUGCCGCAUCAUAAAGCAGCCGAGGAGCCAUGCUCUGCUCAUCGGGGUCGGAGGAUCCGGACGCCAGAGCUUGACUCGAUUGGCUUCGCAUAUAUGCGACU ..........((((((.((((((.((........))((((((.(((.((.((((((..(.((.(((((....))))).)).).)))))).)).)))..))))))..)))))).)))))). ( -50.20) >consensus UCGAGCAUUUAUCGCGCAUAUGCCGCAUCAUAAAGCAGCCAAGGAGUCAUGCUCUACUCAUCGGGGUCGGUGGCUCCGGACGCCAGAGCUUGACUCGAUUGGCUUCGCAUAUAUGCGACU ...........(((((.((((((.((........))(((((...(((((.(((((.....(((((((.....))))))).....))))).)))))....)))))..)))))).))))).. (-40.77 = -40.72 + -0.06)
| Location | 17,507,029 – 17,507,149 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -38.64 |
| Energy contribution | -38.17 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
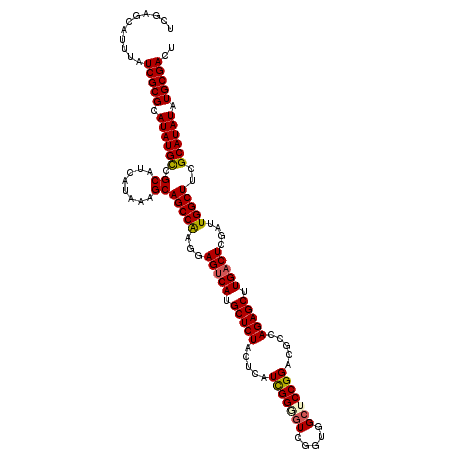
>2L_DroMel_CAF1 17507029 120 - 22407834 AGUCGCAUAUAUGCGAAGCCAAUCGAGUCAAGCUCUGGCGUCCGGAGCCACCAACCCCGAUGAGUAGAGCAUGACUCCUCGGCUGCUUUAUGAUGCGGCAUAUGCGCGACAAAUGCUCGA .(((((.(((((((..((((....((((((.((((((.((((.((..........)).))))..)))))).))))))...))))((........)).))))))).))))).......... ( -49.00) >DroVir_CAF1 27739 120 - 1 AGUCGCAUAUAUGCGAAGCCAAACGAGUCAAGCUCUGACGCCCGGAGCCGCCGACACCGAUGAGCAGAGCAUGACUACGUGGCUGCUUUAUGAUGCGACAUAUGCGCGAUAAAUGUUCGA .((((((((((.((..(((((....(((((.((((((.....(((.(.......).))).....)))))).)))))...)))))))..))).))))))).......(((.......))). ( -42.70) >DroGri_CAF1 32059 120 - 1 AGUCGCAUAUAUGCGAAGCCAGUCGAGUCAAGCUCUGGCGCCCGGAGCCACCGACACCGAUGAGCAGAGCAUGACUACGUGGCUGCUUUAUGAUGCGACAUAUGCGCGAUAAAUGCUCGA .(((((.(((((((..(((((..(((((((.((((((.((..(((.(.......).))).))..)))))).))))).)))))))((........))).)))))).))))).......... ( -41.60) >DroEre_CAF1 24413 120 - 1 AGUCGCAUAUAUGCGAAGCCAAUCGAGUCAAGCUCUGGCGCCCGGAGCCUCCAACUCCGAUAAGUAGAGCAUGACUCCUCGGCUGCUUUAUGAUGCGGCAUAUGCGCGACAAAUGCUCGA .(((((.(((((((..((((....((((((.((((((.(...(((((.......)))))....))))))).))))))...))))((........)).))))))).))))).......... ( -50.00) >DroWil_CAF1 70348 120 - 1 AGUCGCAUAUAUGCGAAGCCAAACGAGUCAAGCUCUGACGUCCGGAGCCACCGACCCCAAUGAGUAGAGCAUGACUACGUGGCUGCUUUAUGAUGCGACAUAUGCGCGAUAAAUGUUCAA .(((((.(((((((..(((((....(((((.((((((.(((..((..(....)..))..)))..)))))).)))))...)))))((........))).)))))).))))).......... ( -39.70) >DroAna_CAF1 25151 120 - 1 AGUCGCAUAUAUGCGAAGCCAAUCGAGUCAAGCUCUGGCGUCCGGAUCCUCCGACCCCGAUGAGCAGAGCAUGGCUCCUCGGCUGCUUUAUGAUGCGGCAUAUGCGCGACAAAUGCUCAA .(((((.(((((((..((((....((((((.((((((.((((.((.((....)).)).))))..)))))).))))))...))))((........)).))))))).))))).......... ( -51.90) >consensus AGUCGCAUAUAUGCGAAGCCAAUCGAGUCAAGCUCUGGCGCCCGGAGCCACCGACCCCGAUGAGCAGAGCAUGACUACGCGGCUGCUUUAUGAUGCGACAUAUGCGCGACAAAUGCUCGA .(((((.(((((((..((((.....(((((.((((((.(...(((...........)))....))))))).)))))....))))((........))).)))))).))))).......... (-38.64 = -38.17 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:13 2006