| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,485,585 – 17,485,695 |
| Length | 110 |
| Max. P | 0.619108 |

| Location | 17,485,585 – 17,485,695 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
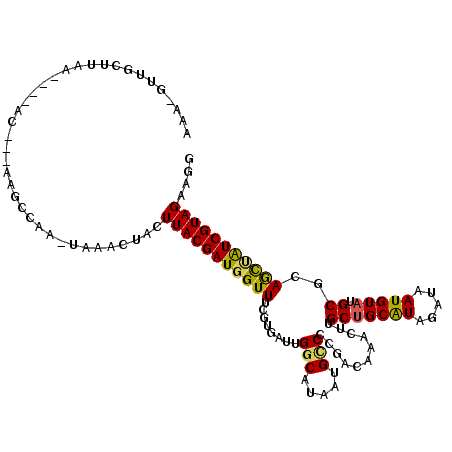
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -18.20 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
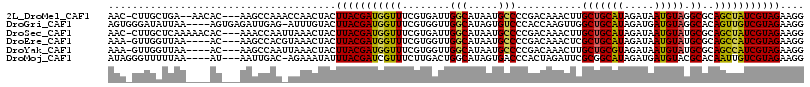
>2L_DroMel_CAF1 17485585 110 + 22407834 AAC-CUUGCUGA--AACAC---AAGCCAAACCAACUACUUACGAUGGUUUCGUGAUUGGCAUAAUGCCCCGACAAACUUGCUGCAUAGAUAAUGUAGGCGCAGCUAUCGUAGAAGG ..(-((((((..--.....---.)))............(((((((((((.(((....(((.....)))............((((((.....))))))))).))))))))))))))) ( -30.20) >DroGri_CAF1 3531 111 + 1 AGUGGGAUAUUAA----AGUGAGAUUGAG-AUUUGUACUUACGAUGGUUUCGUGGUUGGCAUAGUGUCCCACCAAGUUGGCUGCAUAGAUGAUGUAGGCACAGUUGUCGUAGAAGG .(((((((((((.----.((.((..((((-((((((....))))..))))))...)).)).)))))))))))(((.(((.((((((.....)))))).)).).))).......... ( -30.90) >DroSec_CAF1 2514 112 + 1 AAC-CUUGCUCAAAAACAC---AAACCAAUUAAACUACUUACGAUGGUUUCGUGAUUGGCAUAAUGCCCCGACAAACUUGCUGCAUAGAUAAUGUAUGCGCAGCUAUCGUAGAAGG ...-...............---............((.((.((((((((.(((.(...(((.....)))))))......(((.(((((.......)))))))))))))))))).)). ( -22.40) >DroEre_CAF1 2524 108 + 1 AAA-GUUGGUUAA----AC---AAGCCACGUAAACUACUUACGAUGGUUUCGUGGUUGGCAUAAUGCCCCGACAAACUCGCUGCAUAGAUAAUGUAUGCGCAGCCAUCGUAGAAGG ...-(((((....----..---.(((((((.((((((.......)))))))))))))(((.....))))))))...((((((((((.....))))).))).))............. ( -30.90) >DroYak_CAF1 2554 108 + 1 AAA-GUUGGUUAA----AC---AAGCCAAUUAAACUACUUACGAUGGUUUCGUGGUUGGCAUAAUGCCCCGACAAACUUGCUGCGUAGAUAAUGUAUGCGCAGCCAUCGUAGAAGG ..(-(((((((..----..---.))))))))...((.((.((((((((((..(((..(((.....))))))..))))..((((((((.........)))))))))))))))).)). ( -36.30) >DroMoj_CAF1 3415 108 + 1 AUAGGGUUUUUAA----AU---AAUUGAC-AGAAAUAUUUACGAUCGUUUCUUGACUGGCAUAGUGACCCACUAGAUUCGCGGCAUAGAUGAUGUACGCACAAUUGUCGUAGAAGG ......((((((.----((---(((((..-............(((((((....))).....(((((...))))))))).(((((((.....)))).))).)))))))..)))))). ( -17.50) >consensus AAA_GUUGCUUAA____AC___AAGCCAA_UAAACUACUUACGAUGGUUUCGUGAUUGGCAUAAUGCCCCGACAAACUUGCUGCAUAGAUAAUGUAUGCGCAGCUAUCGUAGAAGG ......................................(((((((((((........(((.....)))...........(((((((.....))))).))..))))))))))).... (-18.20 = -17.70 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:09 2006