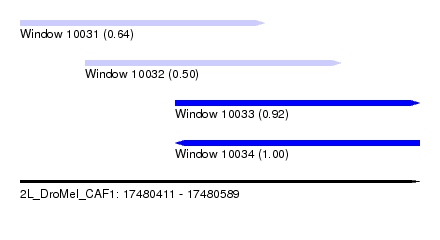
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,480,411 – 17,480,589 |
| Length | 178 |
| Max. P | 0.998822 |

| Location | 17,480,411 – 17,480,520 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -25.23 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17480411 109 + 22407834 -----------UGACCAACUGUCUUUUCGGCAGUGUGGCAACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUGGAGGAUCAUAGAGCCCAUAAAAUAAUGGAGUUCCUGAC -----------(((((.(((......((((((((.(((((....))))).....))))))))((((((....)))))))))...)).)))..((((((((......)))).))))..... ( -34.40) >DroSim_CAF1 1 109 + 1 -----------UGAACCACUGUCUUUUCAGCAAUGUGGCAACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGAUCAAAUAGCCCAUAAAACAAUGGAGUUCCUGAC -----------(((.((.(((.(((((.(((((..(((((..((((......)))))))))..))))).......)))))))).)).)))...(((((((......)))).)))...... ( -24.20) >DroYak_CAF1 81 120 + 1 CCAAGCACUGUUAACCCAUAUUCUAUUCAGCAAUGUGGCAACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGACCAAAGAGCCCAUGAAAUAAUGGAGUUCCUGAC ....(((.((((...(((((((((....)).))))))).)))).)))....((((.....((((..((((((.....))))))..))))...((((((((......)))).)))))))). ( -30.60) >consensus ___________UGACCCACUGUCUUUUCAGCAAUGUGGCAACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGAUCAAAGAGCCCAUAAAAUAAUGGAGUUCCUGAC ..........................((((......((((..((((......))))))))((((..((((((.....))))))..))))...((((((((......)))).)))))))). (-25.23 = -24.90 + -0.33)


| Location | 17,480,440 – 17,480,554 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -29.45 |
| Energy contribution | -28.79 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17480440 114 + 22407834 ACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUGGAGGAUCAUAGAGCCCAUAAAAUAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACAGCGCCCGAG .((((((((.....(((.(((.((((((....))))))..))).((.(((..((((((((......)))).)))).)))))..))).)))(((.....)))..)))))...... ( -30.10) >DroSim_CAF1 30 114 + 1 ACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGAUCAAAUAGCCCAUAAAACAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUGCGCCCGAG .(((((((..(((..((((...((((((....)))))).))))..)))......(((((((.......((....)).........)))))))......))))...)))...... ( -30.79) >DroYak_CAF1 121 114 + 1 ACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGACCAAAGAGCCCAUGAAAUAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUACGCCGGUA ..((((......))))((((((((((((....)))))..((((....(((..((((((((......)))).))))((......))..)))(((.....)))..))))))))))) ( -35.00) >consensus ACGCUGCCAAAUCAGCUGCCGGUUUGUUAUUUAACAAAAGUAGAGGAUCAAAGAGCCCAUAAAAUAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUGCGCCCGAG ..((((......))))((((((((..((((((.....))))))..)))).....(((((((.......((....)).........)))))))......))))............ (-29.45 = -28.79 + -0.66)

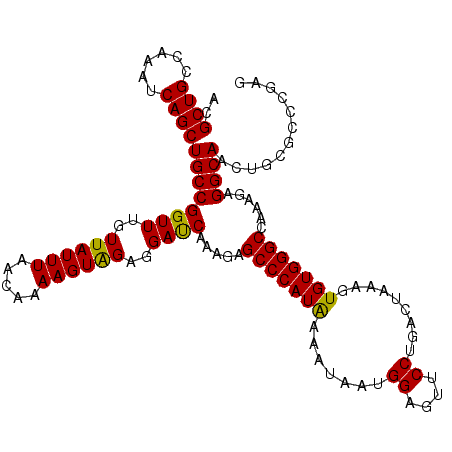
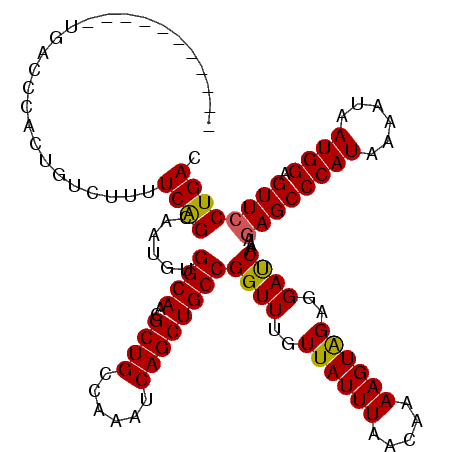
| Location | 17,480,480 – 17,480,589 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -18.28 |
| Energy contribution | -20.28 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
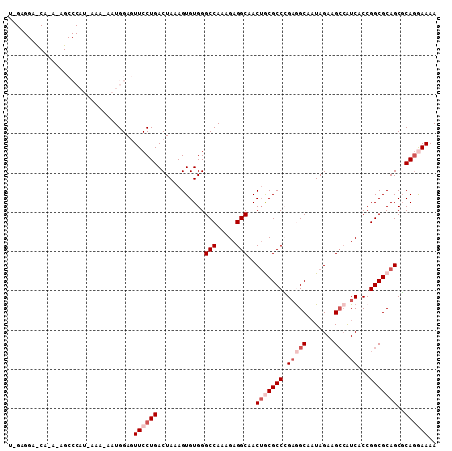
>2L_DroMel_CAF1 17480480 109 + 22407834 UGGAGGAUCAUAGAGCCCAUAAAAUAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACAGCGCCCGAGGCAAUAGAAGCCAUCACCGGCGCAGCGCAAGAAAA ....((.(((..((((((((......)))).)))).)))))...((((..(((.....)))....(((((.(((((.......))).))...))))).))))....... ( -32.00) >DroSec_CAF1 1 83 + 1 --------------------------AUGGAAUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUGCGCCCGAGGCAAUAGAAGACAUCACCGGCGCAGCGCAGGAAAA --------------------------......((((((............(((.....)))..(((((((.((..(.......)...))...)))))))..)))))).. ( -24.50) >DroSim_CAF1 70 109 + 1 UAGAGGAUCAAAUAGCCCAUAAAACAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUGCGCCCGAGGCAAUAGAAGCCAUCACCGGCGCAGCGCAGGAAAA ................((((......))))..((((((............(((.....)))..(((((((.(((((.......))).))...)))))))..)))))).. ( -33.20) >DroEre_CAF1 1 83 + 1 --------------------------AUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUACGCCGGAAGCAUUAGAAGCCGUCACCGGCGCAGCUCAGAAAAA --------------------------...(((((.(...((((.(((((((((.....)))..))))))(....)..))))..((((....))))).)))))....... ( -23.70) >DroYak_CAF1 161 109 + 1 UAGAGGACCAAAGAGCCCAUGAAAUAAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUACGCCGGUAGCAAUAGAAGCAGUCACCGGCGCAGCCCAGAAAAA ............((((((((......)))).))))..........(.(((((.....((....)).(((((((..(..........)..)))))))..))))).).... ( -33.80) >consensus U_GAGGA_CA_A_AGCCCAU_AAA_AAUGGAGUUCCUGACUAAAGUGUGGGCCAAAGAGGCAACUGCGCCCGAGGCAAUAGAAGCCAUCACCGGCGCAGCGCAGGAAAA ................................((((((............(((.....)))..(((((((.(((((.......))).))...)))))))..)))))).. (-18.28 = -20.28 + 2.00)


| Location | 17,480,480 – 17,480,589 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
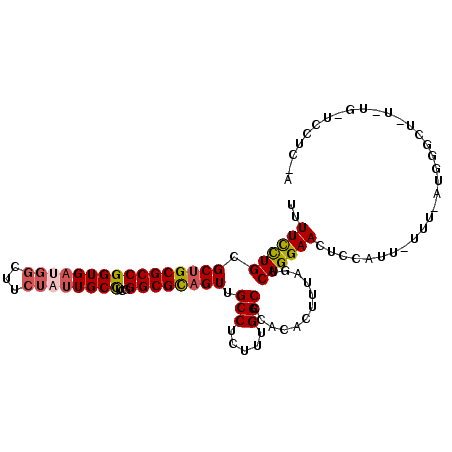
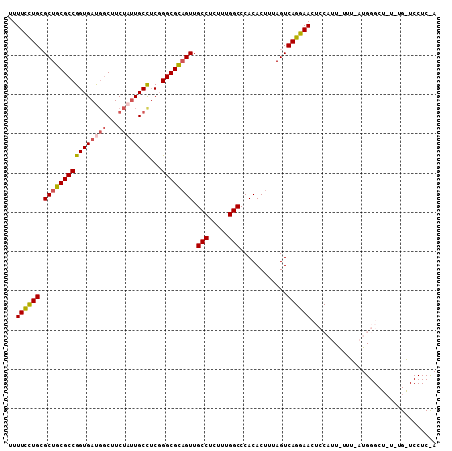
>2L_DroMel_CAF1 17480480 109 - 22407834 UUUUCUUGCGCUGCGCCGGUGAUGGCUUCUAUUGCCUCGGGCGCUGUUGCCUCUUUGGCCCACACUUUAGUCAGGAACUCCAUUAUUUUAUGGGCUCUAUGAUCCUCCA ..((((((.((((((((((..((((...))))..))...)))))(((.(((.....)))..)))....))))))))).(((((......)))))............... ( -30.60) >DroSec_CAF1 1 83 - 1 UUUUCCUGCGCUGCGCCGGUGAUGUCUUCUAUUGCCUCGGGCGCAGUUGCCUCUUUGGCCCACACUUUAGUCAGGAAUUCCAU-------------------------- ..(((((((((((((((((..(((.....)))..))...)))))))).(((.....)))..........).))))))......-------------------------- ( -31.80) >DroSim_CAF1 70 109 - 1 UUUUCCUGCGCUGCGCCGGUGAUGGCUUCUAUUGCCUCGGGCGCAGUUGCCUCUUUGGCCCACACUUUAGUCAGGAACUCCAUUGUUUUAUGGGCUAUUUGAUCCUCUA ..(((((((((((((((((..((((...))))..))...)))))))).(((.....)))..........).)))))).(((((......)))))............... ( -36.90) >DroEre_CAF1 1 83 - 1 UUUUUCUGAGCUGCGCCGGUGACGGCUUCUAAUGCUUCCGGCGUAGUUGCCUCUUUGGCCCACACUUUAGUCAGGAACUCCAU-------------------------- ..(((((((((((((((((....(((.......))).)))))))))).(((.....)))...........)))))))......-------------------------- ( -31.40) >DroYak_CAF1 161 109 - 1 UUUUUCUGGGCUGCGCCGGUGACUGCUUCUAUUGCUACCGGCGUAGUUGCCUCUUUGGCCCACACUUUAGUCAGGAACUCCAUUAUUUCAUGGGCUCUUUGGUCCUCUA ..(((((((((((((((((((...((.......)))))))))))))))(((.....)))............))))))..............(((((....))))).... ( -36.80) >consensus UUUUCCUGCGCUGCGCCGGUGAUGGCUUCUAUUGCCUCGGGCGCAGUUGCCUCUUUGGCCCACACUUUAGUCAGGAACUCCAUU_UUU_AUGGGCU_U_UG_UCCUC_A ..((((((.((((((((((((((((...))))))))...)))))))).(((.....)))............))))))................................ (-25.98 = -26.30 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:07 2006