
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,456,934 – 17,457,042 |
| Length | 108 |
| Max. P | 0.931513 |

| Location | 17,456,934 – 17,457,042 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.47 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -14.86 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17456934 108 + 22407834 CUUCAUCAUCUAUCUGAAUCGUCUUUGAGACGUAGCCUGGUGCUACUUU-AGGUUCAAUGGCUGCAAGCCAUGCGGGUAAUAUACAUUUCGA-----------GCAAAAAUCUGAUGACU ..((((((..(((((((((((((.....)))(((((.....)))))...-.))))).(((((.....)))))..)))))...........((-----------.......)))))))).. ( -27.70) >DroSec_CAF1 21106 83 + 1 CUUC---AUCUGUCUGAAUCGUCUUUGAGACGUAGCCUGGUGCUACUUU-AGGUUCAAUGGCUGCAAGCCAUGCGGGUAAUAUACAU--------------------------------- ....---((((((.(((((((((.....)))(((((.....)))))...-.))))))(((((.....))))))))))).........--------------------------------- ( -25.50) >DroSim_CAF1 21366 86 + 1 CUUCAUCAUCUUUCUGAAUCGUCUUUGAGACGUAGCCUGGUGCUACUUU-AGGUUCAAUGGCUGCAAGCCAUGCGGGUAAUAUACAU--------------------------------- .......((((...(((((((((.....)))(((((.....)))))...-.))))))(((((.....)))))..)))).........--------------------------------- ( -21.70) >DroEre_CAF1 22620 105 + 1 CUUCAUCAUCAUUCAGAAUCGUCUUUGAGACGUAGCCU-GUGCUACUCU-AGAUUCAAUGGCUGCGAGCCAUG--GCUAAUAUACAUUUCGA-----------AAAAGAAUCUGAUCUCU ............(((((.((...(((((((((((((((-(..(((...)-))...))..)))))))(((....--)))........))))))-----------)...)).)))))..... ( -27.20) >DroYak_CAF1 24155 91 + 1 CUUCAUCAUCAUUCAGAAUCGUCUUUGAGACGUAGCCUGGUGCUACUCU-AGGUUCAAUGGCUGCAAG----------------CAUUUCGA-----------GCAAAA-ACUGAUCUCU ............((((....(((.....)))((((((..(.(((.....-.))).)...))))))..(----------------(.......-----------))....-.))))..... ( -19.50) >DroAna_CAF1 16259 113 + 1 CAUC---UUCAUUUAGAAUCGUCUUAGAGACGUAGCCACUUGCUACACACAGACCUGAUACCUGAUUGUAUU----UAAAUAUACAUUUCAGUCCCGAUCUACCAUAGAAUCGGGCUUUG ..((---(......)))((((((.....)))(((((.....)))))..........)))..((((.(((((.----.....)))))..)))).(((((((((...))).))))))..... ( -26.20) >consensus CUUCAUCAUCAUUCAGAAUCGUCUUUGAGACGUAGCCUGGUGCUACUCU_AGGUUCAAUGGCUGCAAGCCAUG__GGUAAUAUACAUUUCGA_____________AA_A_UCUGAU_UCU ...............((((((((.....)))(((((.....))))).....))))).(((((.....)))))................................................ (-14.86 = -16.00 + 1.14)

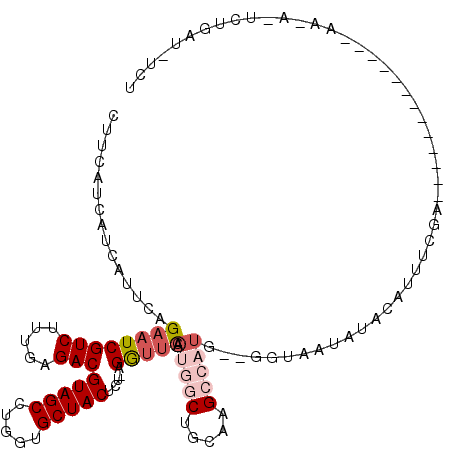

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:58 2006