
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,451,066 – 17,451,165 |
| Length | 99 |
| Max. P | 0.991467 |

| Location | 17,451,066 – 17,451,165 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.06 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -14.03 |
| Energy contribution | -13.37 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
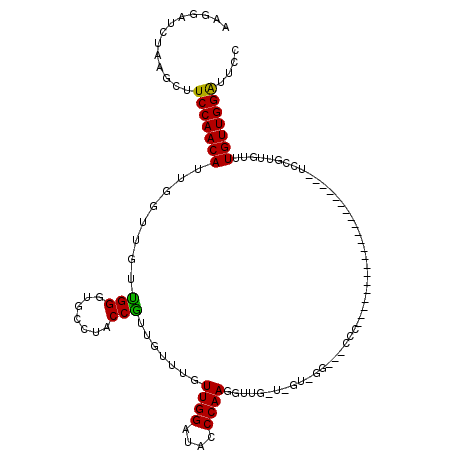
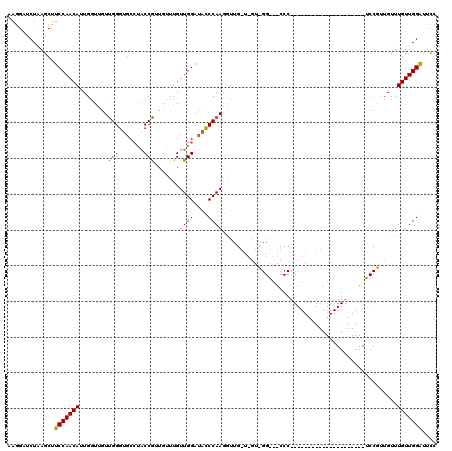
>2L_DroMel_CAF1 17451066 99 + 22407834 AAAGCUCUGAGCUUCCAACAUUGGUAGUCGGGUUCCCUCCGUUGUUUGUUGGAUUUCCAAAAUUGGUAGUCGGGUUCCC---------------------UCCGUUGUUUGUUGGAUUUC ..(((.....)))(((((((......(..(((..(((..(........((((....))))........)..)))..)))---------------------..)......))))))).... ( -27.79) >DroSec_CAF1 15228 78 + 1 AAGGAUCUAAGCUUCCAACAUUGGUUUUUGGGUGCAUACCAUUGUUUGUUGGAUACCCAAGGU------------------------------------------UGUUUGUUGGACGCC ..........((.(((((((.....(((((((((....(((........))).))))))))).------------------------------------------....))))))).)). ( -24.60) >DroSim_CAF1 15425 120 + 1 AAGGAUCUAAGCUUCCAACAUUGUUUGUUGGGUGCCUACCGUUGUUGGUUGGAUACCCAAGGUUGUUUGUUGGACGCCCCCCGAAAGACGUUGGAUUCCCUCCGUUGUUUGUUGGGUUCC ..(((.....((.(((((((.....(.(((((((((.((((....)))).)).))))))).).....))))))).))..(((((.(((((.((((.....)))).))))).))))).))) ( -46.00) >consensus AAGGAUCUAAGCUUCCAACAUUGGUUGUUGGGUGCCUACCGUUGUUUGUUGGAUACCCAAGGUUG_U_GU_GG___CCC_____________________UCCGUUGUUUGUUGGAUUCC .............(((((((........(((.......))).......((((....)))).................................................))))))).... (-14.03 = -13.37 + -0.66)

| Location | 17,451,066 – 17,451,165 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.06 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -10.93 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
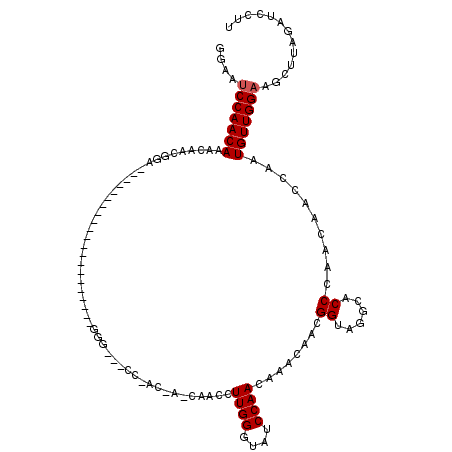
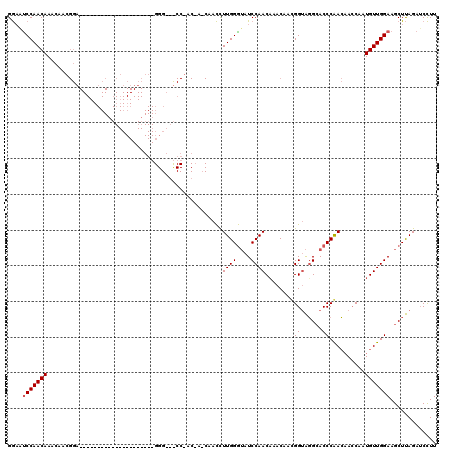
>2L_DroMel_CAF1 17451066 99 - 22407834 GAAAUCCAACAAACAACGGA---------------------GGGAACCCGACUACCAAUUUUGGAAAUCCAACAAACAACGGAGGGAACCCGACUACCAAUGUUGGAAGCUCAGAGCUUU ....(((((((......(..---------------------(((..(((..(........((((....))))........)..)))..)))..)......)))))))(((.....))).. ( -25.89) >DroSec_CAF1 15228 78 - 1 GGCGUCCAACAAACA------------------------------------------ACCUUGGGUAUCCAACAAACAAUGGUAUGCACCCAAAAACCAAUGUUGGAAGCUUAGAUCCUU (((.(((((((....------------------------------------------...((((((..(((........))).....)))))).......))))))).)))......... ( -20.94) >DroSim_CAF1 15425 120 - 1 GGAACCCAACAAACAACGGAGGGAAUCCAACGUCUUUCGGGGGGCGUCCAACAAACAACCUUGGGUAUCCAACCAACAACGGUAGGCACCCAACAAACAAUGUUGGAAGCUUAGAUCCUU (((.(((..........(((.....)))..((.....)))))(((.(((((((.......((((((..((.(((......))).)).)))))).......))))))).)))....))).. ( -37.44) >consensus GGAAUCCAACAAACAACGGA_____________________GGG___CC_AC_A_CAACCUUGGGUAUCCAACAAACAACGGUAGGCACCCAACAACCAAUGUUGGAAGCUUAGAUCCUU ....(((((((.................................................((((....))))........((......))..........)))))))............. (-10.93 = -11.27 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:57 2006