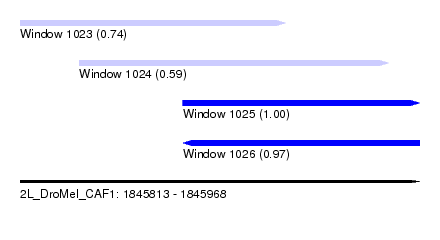
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,845,813 – 1,845,968 |
| Length | 155 |
| Max. P | 0.996698 |

| Location | 1,845,813 – 1,845,916 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.97 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -9.16 |
| Energy contribution | -8.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1845813 103 + 22407834 -----------------UCCAUUUGAACUUUUGAAAAAAGCAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGA -----------------....................((((........((((........((((((.....))))))...))))..((((((....))))))..........))))... ( -16.80) >DroSec_CAF1 13960 103 + 1 -----------------UCCAUUUGAAUUUCUUAAAAAAUCAGUUGUAGCAGAUUUAAAAUCCAUUAAUAAUUUGUGGCCCUCUGAAAACCACAAAUGUGGUUAUUAAUUUAAGCUUAGA -----------------.............((((((...((((..((.(((((((..............))))))).))...)))).((((((....)))))).....))))))...... ( -18.84) >DroSim_CAF1 14072 103 + 1 -----------------UCCAUUUGAACUUCUUAAAAAAUCAGUUGUAGCUGAUUUAAAAUCCAUUAAUAAUUUGUGGCCUUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGA -----------------..........((.((((((((((((((....)))))))).....(((((.....(((((((...........)))))))))))).......))))))...)). ( -19.70) >DroEre_CAF1 14103 120 + 1 GAACUUAAUGUUUCUUUUGAAGUUUAAUUUUUAUAAUAAGCAAUAACGGGAGAUGCAAAAUGUAUAAAUAAUUUGCGGUCCUUUAAUAGCCACAGGAGUGGUUAUUUAAUUAAGUUUAGA (((((((((((((.((.(((((......))))).)).))))......((((..((((((.(((....))).)))))).))))..(((((((((....)))))))))..)))))))))... ( -28.70) >DroYak_CAF1 14354 120 + 1 GAGCUUAAUAUUUUGUUUAAAGCUUAAUUUGAUUAAUAAAAAAUAACAGGAGUUGUAAAGCGCUUAAAUUAUUUGUGGUCUUAUGAUAACCGCGAAAGUGGUAAUUAAUUUAAGUUUAGA ..((((....(((((((((((......)))))...)))))).(((((....))))).))))((((((((((......(((....))).(((((....)))))...))))))))))..... ( -25.20) >consensus _________________UCCAUUUGAAUUUCUUAAAAAAGCAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGA .............................................((.(((((((..............))))))).))....(((.((((((....)))))).)))............. ( -9.16 = -8.88 + -0.28)

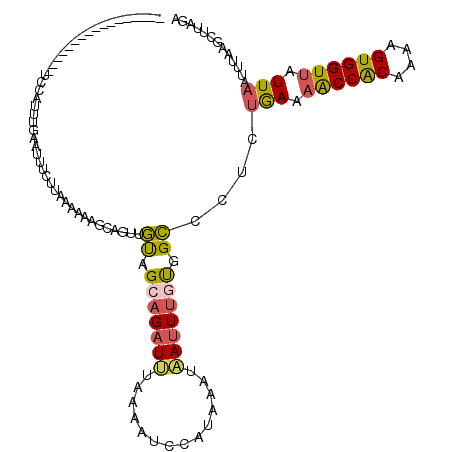
| Location | 1,845,836 – 1,845,956 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1845836 120 + 22407834 CAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUU ..(((((((((((........((((((.....))))))...))))..((((((....)))))).........((((((............))))))..)))))))............... ( -22.90) >DroSec_CAF1 13983 119 + 1 CAGUUGUAGCAGAUUUAAAAUCCAUUAAUAAUUUGUGGCCCUCUGAAAACCACAAAUGUGGUUAUUAAUUUAAGCUUAGAAAA-AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUU ..(((((((((((((......((((.....((((((((...........)))))))))))).....))))).((((((.....-......))))))).)))))))............... ( -21.80) >DroSim_CAF1 14095 119 + 1 CAGUUGUAGCUGAUUUAAAAUCCAUUAAUAAUUUGUGGCCUUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAA-AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUU ..(((((((.(((.((((...(((((.....(((((((...........))))))))))))...)))).)))((((((.....-......))))))..)))))))............... ( -21.80) >DroEre_CAF1 14143 119 + 1 CAAUAACGGGAGAUGCAAAAUGUAUAAAUAAUUUGCGGUCCUUUAAUAGCCACAGGAGUGGUUAUUUAAUUAAGUUUAGAAAA-AAAGCACAAGCUGAUUAUAACCUAUUAACCAUUAUU .(((((.((..((((((((.(((....))).)))))(((.(((.(((((((((....))))))))).....))).........-..(((....))).......))).)))..)).))))) ( -22.90) >DroYak_CAF1 14394 119 + 1 AAAUAACAGGAGUUGUAAAGCGCUUAAAUUAUUUGUGGUCUUAUGAUAACCGCGAAAGUGGUAAUUAAUUUAAGUUUAGAAAA-AAAGCAUAAGCUGAUUAUAACCUAAUGCCCAUUAUU ........((.(((((((...((((((((((......(((....))).(((((....)))))...))))))))))........-..(((....)))..)))))))))............. ( -25.50) >consensus CAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAA_AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUU ...............................(((((((...........)))))))((((((((........((((((............)))))).............))))))))... (-13.98 = -14.14 + 0.16)

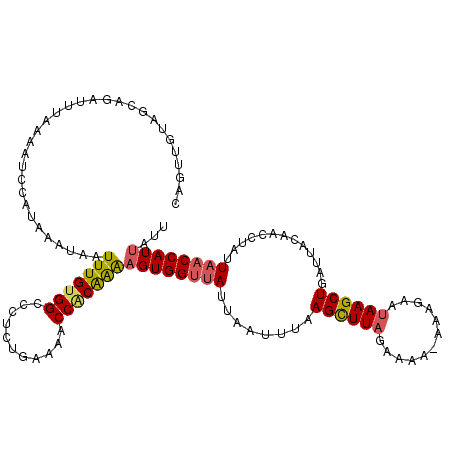
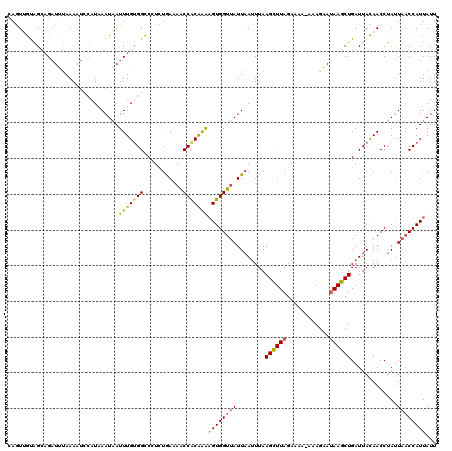
| Location | 1,845,876 – 1,845,968 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1845876 92 + 22407834 CUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUUUGCGGUUACUAU .......((((.((((((((((((.(((((..((((((............)))))))))))........)))))))).)))).))))..... ( -19.00) >DroSec_CAF1 14023 91 + 1 CUCUGAAAACCACAAAUGUGGUUAUUAAUUUAAGCUUAGAAAA-AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUUUGCGGUUACUAU .......((((.((((((((((((.(((((..((((((.....-......)))))))))))........)))))).)))))).))))..... ( -19.70) >DroSim_CAF1 14135 91 + 1 UUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAA-AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUUUGCGGUUACUAC .......((((.((((((((((((.(((((..((((((.....-......)))))))))))........)))))))).)))).))))..... ( -19.10) >DroEre_CAF1 14183 91 + 1 CUUUAAUAGCCACAGGAGUGGUUAUUUAAUUAAGUUUAGAAAA-AAAGCACAAGCUGAUUAUAACCUAUUAACCAUUAUUUGCGGUUAUUUA ....(((((((.(((((((((((((((((......)))))...-..(((....))).............)))))))).)))).))))))).. ( -20.40) >DroYak_CAF1 14434 91 + 1 UUAUGAUAACCGCGAAAGUGGUAAUUAAUUUAAGUUUAGAAAA-AAAGCAUAAGCUGAUUAUAACCUAAUGCCCAUUAUUUGCGGUUAUUUA ....(((((((((((((((((..((((..(((((((((.....-......)))))).....)))..))))..))))).)))))))))))).. ( -23.00) >consensus CUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAA_AAAGAAUAAGCUGAUUACAACCUAUUAACCAUUAUUUGCGGUUACUAA .......((((.((((((((((((........((((((............)))))).............)))))))).)))).))))..... (-15.90 = -15.82 + -0.08)

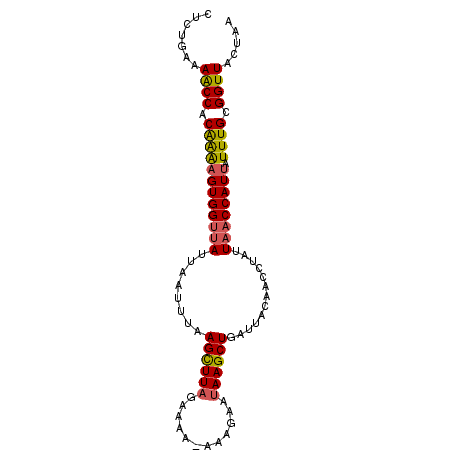
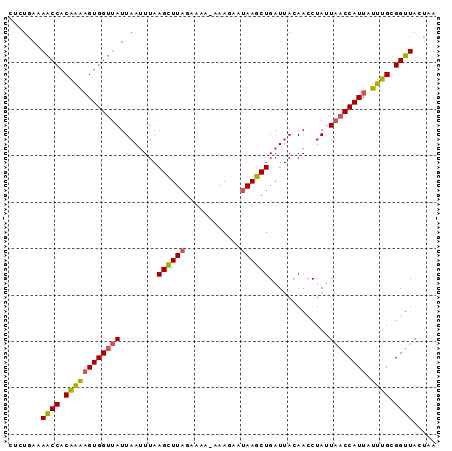
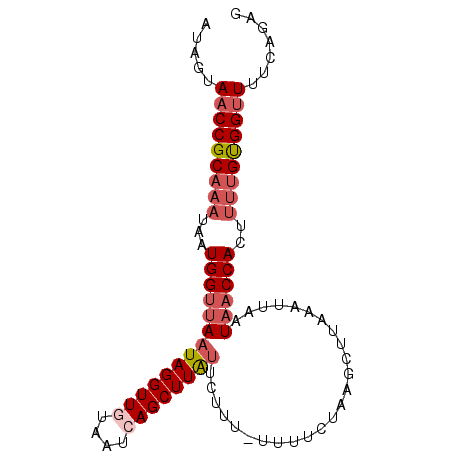
| Location | 1,845,876 – 1,845,968 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -14.95 |
| Energy contribution | -16.43 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1845876 92 - 22407834 AUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUCAGAG ...(.(((((((((...((((((((((((((....)))))))).(((........)))..........))))))..)))))))))..).... ( -21.70) >DroSec_CAF1 14023 91 - 1 AUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUU-UUUUCUAAGCUUAAAUUAAUAACCACAUUUGUGGUUUUCAGAG ...(.((((((((((..((((((((((((((....)))))))).(((.-......)))..........)))))).))))))))))..).... ( -21.70) >DroSim_CAF1 14135 91 - 1 GUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUU-UUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUUAGAA .(((.(((((((((...((((((((((((((....)))))))).(((.-......)))..........))))))..))))))))).)))... ( -22.20) >DroEre_CAF1 14183 91 - 1 UAAAUAACCGCAAAUAAUGGUUAAUAGGUUAUAAUCAGCUUGUGCUUU-UUUUCUAAACUUAAUUAAAUAACCACUCCUGUGGCUAUUAAAG ..((((.(((((.....((((((.((((((..((..(((....)))..-)).....))))))......))))))....))))).)))).... ( -15.20) >DroYak_CAF1 14434 91 - 1 UAAAUAACCGCAAAUAAUGGGCAUUAGGUUAUAAUCAGCUUAUGCUUU-UUUUCUAAACUUAAAUUAAUUACCACUUUCGCGGUUAUCAUAA ...((((((((.((...(((((((.(((((......)))))))))(((-......))).............)))..)).))))))))..... ( -17.20) >consensus AUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUU_UUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUCAGAG .....(((((((((...((((((((((((((....)))))))).........................))))))..)))))))))....... (-14.95 = -16.43 + 1.48)

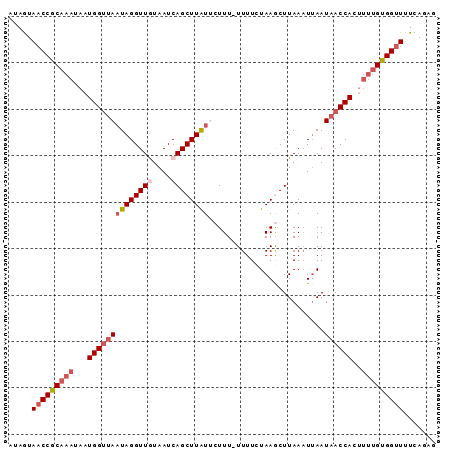
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:21 2006