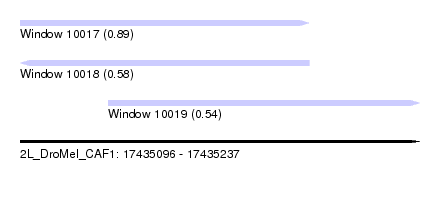
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,435,096 – 17,435,237 |
| Length | 141 |
| Max. P | 0.892239 |

| Location | 17,435,096 – 17,435,198 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
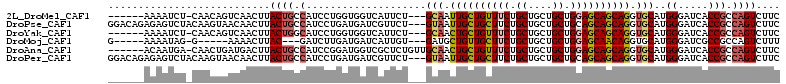
>2L_DroMel_CAF1 17435096 102 + 22407834 ------AAAAUCU-CAACAGUCAACUUACUGCCAUCCUGGUGGUCAUUCU---GCAAUUGCUGUUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ------.......-...((((......)))).....((((((((.(((((---(((.((((((((((.((....)).)))))))))).)))..))))).))))))))..... ( -40.10) >DroPse_CAF1 32621 109 + 1 GGACAGAGAGUCUACAAGUAACAACUUACUGCCAUCCUGAUGAUCGUUCU---GUAAUUGCUGCUUCUGCUGCUGCUGCAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ((.(((.((((............)))).)))))...(((.(((((.(..(---(((.((((((((.(.((....)).).)))))))).)))).).)))))....)))..... ( -31.70) >DroYak_CAF1 23418 102 + 1 ------AAAAUCU-CAACAGUCAACUUACUGGCAUCCUGGUGGUCAUUCU---GCAACUGCUGUUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ------.......-.....((((......))))...((((((((.(((((---(((.((((((((((.((....)).)))))))))).)))..))))).))))))))..... ( -43.40) >DroMoj_CAF1 23782 95 + 1 G-----AAAAUAG-G-----AAAACUUAC---GAUCUUGAUGAUCAUUGU---GAUGCUGUUGCUUCUGCUGCUGCUGGAGCAACAGGUGCAUGGGAUCGCCGCCAGUCUUU .-----.....((-(-----(.......(---((((((.((((((.....---))).((((((((((.((....)).))))))))))...)))))))))).......)))). ( -29.34) >DroAna_CAF1 24860 105 + 1 ------ACAAUGA-CAACUGAUGACUUACUGCCAUCCGGAUGGUCGCUCUGUUGCAACUGCUGUUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ------.....((-(....((((.(.....).))))(((.(((((.(.....((((.((((((((((.((....)).)))))))))).)))).).))))))))...)))... ( -39.70) >DroPer_CAF1 32398 109 + 1 GGACAGAGAGUCUACAAGUAACAACUUACUGCCAUCCUGAUGAUCGUUCU---GUAAUUGCUGCUUCUGCUGCUGCUGCAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ((.(((.((((............)))).)))))...(((.(((((.(..(---(((.((((((((.(.((....)).).)))))))).)))).).)))))....)))..... ( -31.70) >consensus ______AAAAUCU_CAACUGACAACUUACUGCCAUCCUGAUGAUCAUUCU___GCAACUGCUGCUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUC ...........................((((.(....................(((.((((((((((.((....)).)))))))))).)))..((.....))).)))).... (-23.71 = -23.93 + 0.22)
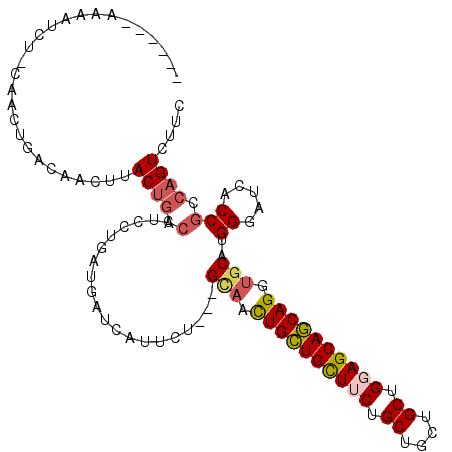
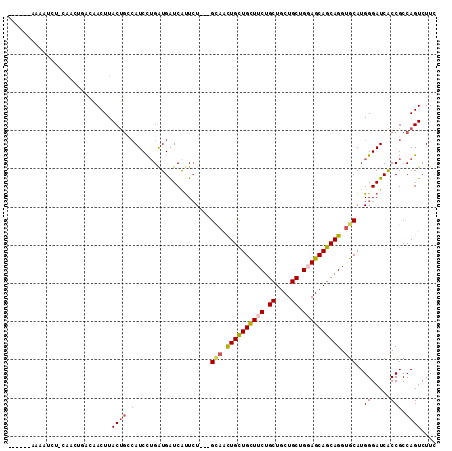
| Location | 17,435,096 – 17,435,198 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17435096 102 - 22407834 GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUCCAGCAGCAGCAGAAACAGCAAUUGC---AGAAUGACCACCAGGAUGGCAGUAAGUUGACUGUUG-AGAUUUU------ .....((((.(((.((....((((..((((((....))))))((.......))...)))---)..)).))).))))...((((((......)))))).-.......------ ( -32.10) >DroPse_CAF1 32621 109 - 1 GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUGCAGCAGCAGCAGAAGCAGCAAUUAC---AGAACGAUCAUCAGGAUGGCAGUAAGUUGUUACUUGUAGACUCUCUGUCC ..........(((((((...(((..(((((((((...)))))))))..)))(......)---.....))))))).....(((((..((((..........))))..))))). ( -33.90) >DroYak_CAF1 23418 102 - 1 GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUCCAGCAGCAGCAGAAACAGCAGUUGC---AGAAUGACCACCAGGAUGCCAGUAAGUUGACUGUUG-AGAUUUU------ ....((((((((((.((...(((..((((((((.....)))))))).(((....)))))---)....)).)))).....)))))).............-.......------ ( -31.60) >DroMoj_CAF1 23782 95 - 1 AAAGACUGGCGGCGAUCCCAUGCACCUGUUGCUCCAGCAGCAGCAGAAGCAACAGCAUC---ACAAUGAUCAUCAAGAUC---GUAAGUUUU-----C-CUAUUUU-----C .((((((....((((((...(((..((((((((.....))))))))..)))...(.(((---.....)))).....))))---)).))))))-----.-.......-----. ( -26.30) >DroAna_CAF1 24860 105 - 1 GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUCCAGCAGCAGCAGAAACAGCAGUUGCAACAGAGCGACCAUCCGGAUGGCAGUAAGUCAUCAGUUG-UCAUUGU------ ((.((((.((.((.((((..(((..((((((((.....)))))))).....)))(((((......))))).....)))).)).)).)))).)).....-.......------ ( -35.90) >DroPer_CAF1 32398 109 - 1 GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUGCAGCAGCAGCAGAAGCAGCAAUUAC---AGAACGAUCAUCAGGAUGGCAGUAAGUUGUUACUUGUAGACUCUCUGUCC ..........(((((((...(((..(((((((((...)))))))))..)))(......)---.....))))))).....(((((..((((..........))))..))))). ( -33.90) >consensus GAAGACUGGCGGUGAUCCCAUGCACCUGCUGCUCCAGCAGCAGCAGAAACAGCAAUUGC___AGAACGACCAUCAGGAUGGCAGUAAGUUGUCAGUUG_AGAUUUU______ ...((((.((.((.((((..(((..((((((((.....)))))))).....))).............(.....).)))).)).)).))))...................... (-21.38 = -21.93 + 0.56)

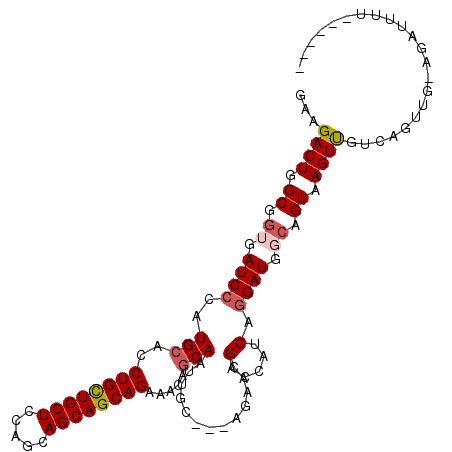
| Location | 17,435,127 – 17,435,237 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -30.87 |
| Energy contribution | -29.65 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17435127 110 + 22407834 GGUGGUCAUUCU---GCAAUU------GCUGUUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUCUGACGUUUCCGCCUCAAGUGCGCUGGAUCUAUAA-GACAA ((((((.(((((---(((.((------((((((((.((....)).)))))))))).)))..))))).)))))).(((((..((....((((((......).)).)))))...))-))).. ( -43.60) >DroPse_CAF1 32659 111 + 1 GAUGAUCGUUCU---GUAAUU------GCUGCUUCUGCUGCUGCUGCAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUCUGACGUUUCCGCCUCAGGUGUGCUGGAUCUACAAAAACAA (.(((((.(..(---(((.((------((((((.(.((....)).).)))))))).)))).).))))).).(((((..(((((.((....)).)))))...))))).............. ( -36.70) >DroGri_CAF1 29095 116 + 1 GAUGAUCAUUCU---GAUGCUGCUGUUGUUGUUUCUGUUGUUGCUGGAGCAACAAGUGCAUUGGAUCACCGCCAGUCUUUUGACGUUUACGCCUCAAAUGUGCUGGAUCUACAA-UAAAA ...((((..((.---.((((.(((..(((((((((.((....)).))))))))))))))))..)).......((((..(((((.((....)).)))))...)))))))).....-..... ( -33.40) >DroMoj_CAF1 23806 110 + 1 GAUGAUCAUUGU---GAUGCU------GUUGCUUCUGCUGCUGCUGGAGCAACAGGUGCAUGGGAUCGCCGCCAGUCUUUUGACGUUUCCGCCUCAAGUGUGCUGGAUCUAUAA-AAAAG (((((((.(..(---(...((------((((((((.((....)).))))))))))...))..)))))....(((((..(((((.((....)).)))))...)))))))).....-..... ( -37.60) >DroAna_CAF1 24891 113 + 1 GAUGGUCGCUCUGUUGCAACU------GCUGUUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUCUGACGUUUCCGCUUCAAGUGCGCUGGAUCUAUAA-AACAA ((((((.(.(((..((((.((------((((((((.((....)).)))))))))).))))..))).)))).(((((...((((.((....)).)).))...)))))))).....-..... ( -39.20) >DroPer_CAF1 32436 111 + 1 GAUGAUCGUUCU---GUAAUU------GCUGCUUCUGCUGCUGCUGCAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUCUGACGUUUCCGCCUCAGGUGUGCUGGAUCUACAAAAACAA (.(((((.(..(---(((.((------((((((.(.((....)).).)))))))).)))).).))))).).(((((..(((((.((....)).)))))...))))).............. ( -36.70) >consensus GAUGAUCAUUCU___GAAACU______GCUGCUUCUGCUGCUGCUGGAGCAGCAGGUGCAUGGGAUCACCGCCAGUCUUCUGACGUUUCCGCCUCAAGUGUGCUGGAUCUACAA_AACAA ...((((....................((((((((.((....)).)))))))).(((((((..((.........(((....))).........))..))))))).))))........... (-30.87 = -29.65 + -1.22)
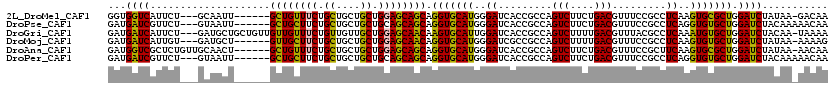
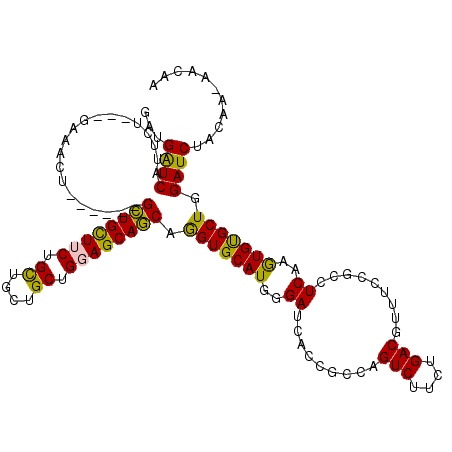
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:53 2006