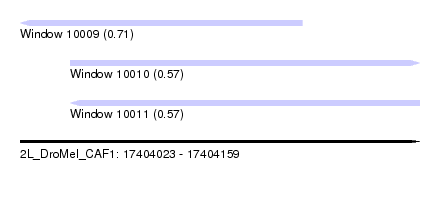
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,404,023 – 17,404,159 |
| Length | 136 |
| Max. P | 0.713522 |

| Location | 17,404,023 – 17,404,119 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -21.54 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17404023 96 - 22407834 ACUCCUGGAGUUCCUUAUUUGUGGCUUGUUGGGCAUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUUUACUAGUUCCUCUACUU .(((((.((((..(......)..))))(((((....))))).)))))..((.((.((((((......))))))))))................... ( -26.60) >DroPse_CAF1 30560 96 - 1 UCUGCAGGAGUUCCUUGUUUUCAGCCUGUUGGGUAUCCAACUGGACCGUGGUUUCAGCUAGUCUUGCCUCCAAUACUUUAACAAGUUCUUUCUUCU ...((((((.(...(((...(((.((.(((((....))))).))....)))...)))..).))))))............................. ( -17.90) >DroSec_CAF1 25869 96 - 1 ACUCCUGGAGUUUCUUAUUUGUGGCUUGUUGGGACUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUUUACUAGUUCCUCAACUU ......(((((..(................)..)))))((((((((((((.....((((((......))))))))))))..))))))......... ( -27.99) >DroSim_CAF1 23675 96 - 1 ACUCCUGGAGUUUCUUAUGUGUGGCUUGUUGGGACUCCAACUGGGAGGUAGCUGCUUGGAGUUUUUCCUCCAACACCUUUACUAGUUCCUCAACUU ...((((((((..(....(.....).....)..))))))...))((((.(((((.((((((......)))))).........)))))))))..... ( -25.50) >DroEre_CAF1 28195 96 - 1 ACUCCCGGAGUUCCCUAUUUUCCGCUUGUUGGGACUCCAACUGGGAGGUGGCGGCUUGGAGCUUUUCCUCUAACACCUUAACUAGUUCCUCCACUU .((((((((((.(((...............))))))))....)))))((((.((.((((((......))))))...((.....))..)).)))).. ( -29.66) >DroYak_CAF1 26457 96 - 1 ACUCCUGGAGUUCCCUAUUUUCCGCUUGCUGGGACUCUAACUGGGAGGUGGCUGCUUGGAGUUUAUCCUCCAACACCUUAACUAGUUCCUCCACUU .....(((((.((((...............))))....((((((((((((.....((((((......))))))))))))..)))))).)))))... ( -28.26) >consensus ACUCCUGGAGUUCCUUAUUUGCGGCUUGUUGGGACUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUUAACUAGUUCCUCCACUU ......((((((((................))))))))((((((((((((.....((((((......))))))))))))..))))))......... (-21.54 = -22.82 + 1.28)

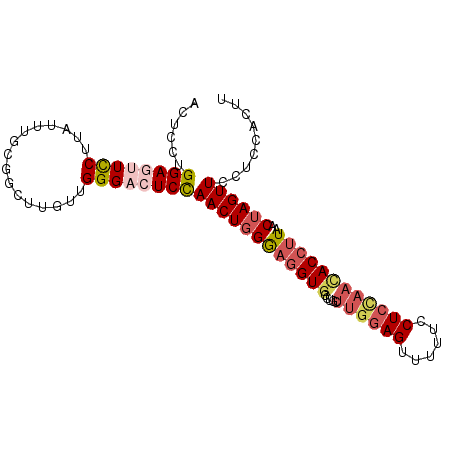
| Location | 17,404,040 – 17,404,159 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -21.85 |
| Energy contribution | -22.93 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17404040 119 + 22407834 AAGGUGUUGGAGGAAAAACUCCAAGCAGCCACCUCCCAGUUGGAUGCCCAACAAGCCACAAAUAAGGAACUCCAGGAGUUGCUGGUUAAAUCUCAAGAAAACGAGGGAAAUCUGCAAGG ..(((.((((((......))))))...)))..((((..(((((....))))).............((....)).))))((((.((((...((((........))))..)))).)))).. ( -35.20) >DroPse_CAF1 30577 119 + 1 AAAGUAUUGGAGGCAAGACUAGCUGAAACCACGGUCCAGUUGGAUACCCAACAGGCUGAAAACAAGGAACUCCUGCAGAAACUCGAACAGACCCAGCAAAAGGAGGGAGUCCUACAGGA ..(((.(((....))).)))..(((......(((((..(((((....))))).)))))......((((.(((((........((.....)).((.......)).))))))))).))).. ( -27.90) >DroSec_CAF1 25886 119 + 1 AAGGUGUUGGAGGAAAAACUCCAAGCAGCCACCUCCCAGUUGGAGUCCCAACAAGCCACAAAUAAGAAACUCCAGGAGUUGCUGGUUAAAUCUCAAGAAAACGAGGGAAAUCUGCAAGG ..(((.((((((......))))))...)))..((((....((((((..(................)..))))))))))((((.((((...((((........))))..)))).)))).. ( -31.89) >DroSim_CAF1 23692 119 + 1 AAGGUGUUGGAGGAAAAACUCCAAGCAGCUACCUCCCAGUUGGAGUCCCAACAAGCCACACAUAAGAAACUCCAGGAGUUGCUGGUUAAAUCUCAAGAAAACGAGGGAAAUCUGCAAGG ......((((((......))))))((((.....((((.(((((....))))).(((((.........(((((...)))))..))))).................))))...)))).... ( -31.40) >DroEre_CAF1 28212 119 + 1 AAGGUGUUAGAGGAAAAGCUCCAAGCCGCCACCUCCCAGUUGGAGUCCCAACAAGCGGAAAAUAGGGAACUCCGGGAGUUGCUGUUAAAAUCGCAAGAGAACGAGGGAAAUCUGCAAGG ..((((.....(((.....)))....))))..(((((....((((((((...............))).))))))))))((((.(((....((....)).)))((......)).)))).. ( -32.36) >DroYak_CAF1 26474 119 + 1 AAGGUGUUGGAGGAUAAACUCCAAGCAGCCACCUCCCAGUUAGAGUCCCAGCAAGCGGAAAAUAGGGAACUCCAGGAGUUGCUGGUGAAAUCUCAAGAGAAAGAGGGAAAUUUACAAGG ..(((.((((((......))))))...)))...((((........(((((((((.(.......((....))......)))))))).))..(((....)))....))))........... ( -32.12) >consensus AAGGUGUUGGAGGAAAAACUCCAAGCAGCCACCUCCCAGUUGGAGUCCCAACAAGCCAAAAAUAAGGAACUCCAGGAGUUGCUGGUUAAAUCUCAAGAAAACGAGGGAAAUCUGCAAGG ..(((.((((((......))))))...)))...((((.(((((....)))))............((.(((((...))))).)).....................))))........... (-21.85 = -22.93 + 1.09)
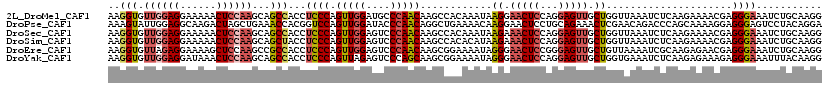
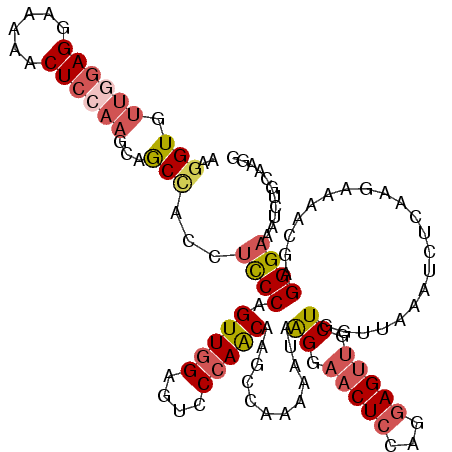
| Location | 17,404,040 – 17,404,159 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17404040 119 - 22407834 CCUUGCAGAUUUCCCUCGUUUUCUUGAGAUUUAACCAGCAACUCCUGGAGUUCCUUAUUUGUGGCUUGUUGGGCAUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUU ....((((......((((......))))......(((....(((((.((((..(......)..))))(((((....))))).))))).)))))))((((((......))))))...... ( -33.40) >DroPse_CAF1 30577 119 - 1 UCCUGUAGGACUCCCUCCUUUUGCUGGGUCUGUUCGAGUUUCUGCAGGAGUUCCUUGUUUUCAGCCUGUUGGGUAUCCAACUGGACCGUGGUUUCAGCUAGUCUUGCCUCCAAUACUUU (((((((((((((...(((......))).......))).))))))))))...............((.(((((....))))).))....(((.....((.......))..)))....... ( -29.20) >DroSec_CAF1 25886 119 - 1 CCUUGCAGAUUUCCCUCGUUUUCUUGAGAUUUAACCAGCAACUCCUGGAGUUUCUUAUUUGUGGCUUGUUGGGACUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUU ....(((((((((((..(((....(((((.....((((......))))....))))).....)))..(((((....))))).)))))))..))))((((((......))))))...... ( -33.70) >DroSim_CAF1 23692 119 - 1 CCUUGCAGAUUUCCCUCGUUUUCUUGAGAUUUAACCAGCAACUCCUGGAGUUUCUUAUGUGUGGCUUGUUGGGACUCCAACUGGGAGGUAGCUGCUUGGAGUUUUUCCUCCAACACCUU ....(((((((((((..(((..(.(((((.....((((......))))....))))).)...)))..(((((....))))).)))))))..))))((((((......))))))...... ( -33.90) >DroEre_CAF1 28212 119 - 1 CCUUGCAGAUUUCCCUCGUUCUCUUGCGAUUUUAACAGCAACUCCCGGAGUUCCCUAUUUUCCGCUUGUUGGGACUCCAACUGGGAGGUGGCGGCUUGGAGCUUUUCCUCUAACACCUU ....((..(((((((((((......)))).................(((((.(((...............))))))))....))))))).))((.((((((......))))))..)).. ( -32.36) >DroYak_CAF1 26474 119 - 1 CCUUGUAAAUUUCCCUCUUUCUCUUGAGAUUUCACCAGCAACUCCUGGAGUUCCCUAUUUUCCGCUUGCUGGGACUCUAACUGGGAGGUGGCUGCUUGGAGUUUAUCCUCCAACACCUU ........(((((((((((......))))..((.(((((((....(((((.........))))).)))))))))........)))))))((.((.((((((......)))))))))).. ( -33.50) >consensus CCUUGCAGAUUUCCCUCGUUUUCUUGAGAUUUAACCAGCAACUCCUGGAGUUCCUUAUUUGCGGCUUGUUGGGACUCCAACUGGGAGGUGGCUGCUUGGAGUUUUUCCUCCAACACCUU ......(((((((............)))))))...((((..(((((((((((((................))))))))....)))))...)))).((((((......))))))...... (-23.05 = -23.47 + 0.43)
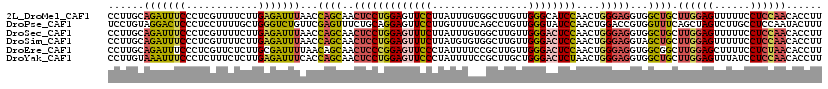
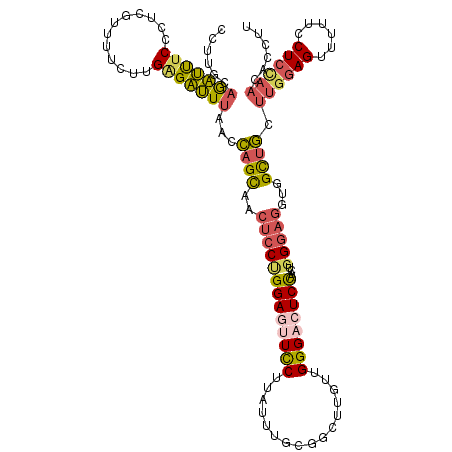
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:45 2006