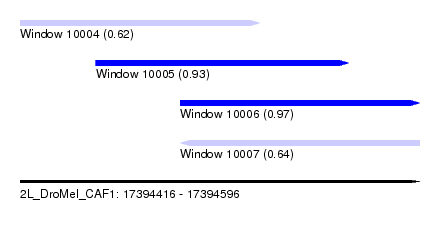
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,394,416 – 17,394,596 |
| Length | 180 |
| Max. P | 0.972771 |

| Location | 17,394,416 – 17,394,524 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17394416 108 + 22407834 CGAA--AUGCUAAACAAUUGC-AAUGCAUUUUAUGGA----AAUUCGGGGCGACUUCGUGUAAAGAUGUAUAUGUAUGUACAUUUUUGUAUAUCUACAAA-CCCGUCCUG---UCUAUA .(((--((((...........-...)))))))(((((----....(((((((....))).((((((((((((....))))))))))))............-)))).....---))))). ( -27.54) >DroSec_CAF1 16394 85 + 1 CGAA--CUGCUAAAAAAUUGC-AAUGCAUUUUAUGGA----AAUUCGGGGCGACU--------------------GUAUACA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUA ....--.(((.........))-)........((((((----....(((((((..(--------------------(....))---(((((....))))).-..)))))))---)))))) ( -15.40) >DroSim_CAF1 15920 101 + 1 CGAA--AUGCUAAACAAUUGC-AAUGCAUUUUAUGGA----AAUUCGGGGCGACUUCGUGUAUA----UAUAUAUAUAUGCA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUA .(((--((((...........-...)))))))(((((----....(((((((......((((((----((....))))))))---(((((....))))).-..)))))))---))))). ( -25.44) >DroEre_CAF1 18390 104 + 1 CGAA--AUGCUAAGCAAUUGCCAAUGCAUUUUAUGGACUCACAUUUGGGGCGACU-CGUGUAUA----UAU--GUAUGUACAU--UUGUAUAUCAACAAA-CCCGUCCUG---UCUAUA .(((--((((...((....))....)))))))((((((........((((((...-.(((((((----(..--..))))))))--((((......)))).-..)))))))---))))). ( -26.50) >DroYak_CAF1 15916 97 + 1 CGAA--AUGCUAAACAAUUGCAAAUGCAUUUAAUGGA----AAUUCGGGGCGACUUCGUGUAUA----UAU------GUACAU--CUGUAUAUCUACAAA-CCCGUCCUG---UCUAUA ..((--((((...............)))))).(((((----....(((((((......((((..----.((------((((..--..)))))).))))..-..)))))))---))))). ( -21.86) >DroAna_CAF1 11422 97 + 1 CCAAAAACGCCAA--AA-UGCGAAUGCAUUUUAUGGA----AAUUCGUCUCGAGUUCUCAUCU----------GUAUGUACAC-----UAUAUAUACAAUAUCUGUGAAAUACGCUAUA .......(((...--..-.)))...((((((((((((----((((((...))))))......(----------(((((((...-----..))))))))...))))))))))..)).... ( -18.10) >consensus CGAA__AUGCUAAACAAUUGC_AAUGCAUUUUAUGGA____AAUUCGGGGCGACUUCGUGUAUA____UAU__GUAUGUACAU__UUGUAUAUCUACAAA_CCCGUCCUG___UCUAUA ..................(((....)))...((((((........(((((((((.....))..............(((((((....)))))))..........)))))))...)))))) ( -9.94 = -10.67 + 0.73)


| Location | 17,394,450 – 17,394,564 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17394450 114 + 22407834 -AAUUCGGGGCGACUUCGUGUAAAGAUGUAUAUGUAUGUACAUUUUUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUU -......((....))((((((((((.......((((((((((....))))))..))))..-..((.((((---...((((.(((((....))))))))))))))))))))))))).... ( -32.70) >DroSec_CAF1 16428 91 + 1 -AAUUCGGGGCGACU--------------------GUAUACA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUU -.....((((((..(--------------------(....))---(((((....))))).-..))))))(---(((((((.(((((....))))))))).))))((.....))...... ( -22.80) >DroSim_CAF1 15954 107 + 1 -AAUUCGGGGCGACUUCGUGUAUA----UAUAUAUAUAUGCA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUU -.....((((((......((((((----((....))))))))---(((((....))))).-..))))))(---(((((((.(((((....))))))))).))))((.....))...... ( -29.60) >DroEre_CAF1 18428 106 + 1 ACAUUUGGGGCGACU-CGUGUAUA----UAU--GUAUGUACAU--UUGUAUAUCAACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUCACGCGAUCUU ......((((((...-.(((((((----(..--..))))))))--((((......)))).-..))))))(---(((((((.(((((....))))))))).))))((.....))...... ( -28.40) >DroYak_CAF1 15951 102 + 1 -AAUUCGGGGCGACUUCGUGUAUA----UAU------GUACAU--CUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUU -.....((((((......((((..----.((------((((..--..)))))).))))..-..))))))(---(((((((.(((((....))))))))).))))((.....))...... ( -27.20) >DroAna_CAF1 11456 103 + 1 -AAUUCGUCUCGAGUUCUCAUCU----------GUAUGUACAC-----UAUAUAUACAAUAUCUGUGAAAUACGCUAUAGAUCUCUAUCAAGAGACCGUCAGACGCUUCACGCGCUCCU -.....((((.(((.((((...(----------(((((((...-----..))))))))..(((((((........))))))).........)))).).))))))((.....))...... ( -21.00) >consensus _AAUUCGGGGCGACUUCGUGUAUA____UAU__GUAUGUACAU__UUGUAUAUCUACAAA_CCCGUCCUG___UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUU .....(((((((((.....))..............(((((((....)))))))..........)))))))....((((((.(((((....))))))))).)).(((.....)))..... (-17.69 = -18.25 + 0.56)


| Location | 17,394,488 – 17,394,596 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
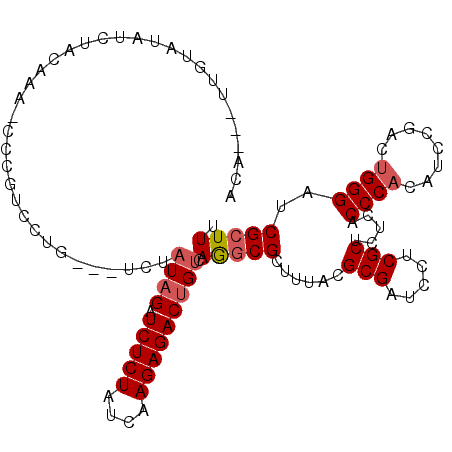
>2L_DroMel_CAF1 17394488 108 + 22407834 ACAUUUUUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACACCCGACUGGGAUCGCUUU .....((((((....))))))-((((((..(---(((((((.(((((....))))))))).)))).......(((.....)))..............))).)))........ ( -26.10) >DroVir_CAF1 15106 86 + 1 A------UCCG----------------A-UA---CUCAUAGAUCUCUAUCAAGAGACUGUCAAGCGCUUUACGCGCUCCUCGCUCUCGCCCUCGUCCGACUGGGAACGCUUC .------(((.----------------.-..---.....((.(((((....)))))))(((.(((((.....)))))....((....))........)))..)))....... ( -18.70) >DroSec_CAF1 16446 105 + 1 ACA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACAUCCGACUGGGAUCGCUUU ...---(((((....))))).-((((((..(---(((((((.(((((....))))))))).)))).......(((.....)))..............))).)))........ ( -25.60) >DroSim_CAF1 15988 105 + 1 GCA---UUGUAUAUCUACAAA-CCCGUCCUG---UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACAUCCGACUGGGAUCGCUUU ((.---(((((....))))).-((((((..(---(((((((.(((((....))))))))).)))).......(((.....)))..............))).)))...))... ( -27.60) >DroWil_CAF1 19763 85 + 1 AAA---U--------UACA----------------AUUUAGAUCUCUAUCAAGAGACUGUAAAGCGCUUUACGCGAUCCUCGCUCUCACCCACAUCCGAUUGGGAUCGCUUU ...---.--------....----------------....((.(((((....)))))))(((((....)))))(((((((.((.((............)).)))))))))... ( -20.20) >DroAna_CAF1 11484 107 + 1 ACAC-----UAUAUAUACAAUAUCUGUGAAAUACGCUAUAGAUCUCUAUCAAGAGACCGUCAGACGCUUCACGCGCUCCUCGCUCUCACCCACAUCCGAUUGGGACCGCUUC .(((-----.((((.....))))..)))......((.((((....))))...((((.((..((.(((.....)))))...)).)))).((((........))))...))... ( -20.40) >consensus ACA___UUGUAUAUCUACAAA_CCCGUCCUG___UCUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCCUCGCUCUCACCCACAUCCGACUGGGAUCGCUUU .....................................((((.(((((....))))))))).(((((......(((.....))).....((((........))))..))))). (-16.69 = -17.13 + 0.44)


| Location | 17,394,488 – 17,394,596 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17394488 108 - 22407834 AAAGCGAUCCCAGUCGGGUGUGGGUGAGAGCGAAGAUCGCGUAAAGCGCCUGACAGUCUCUUGAUAGAGAUCUAUAGA---CAGGACGGG-UUUGUAGAUAUACAAAAAUGU ...((((.(((.(((((((((...((...(((.....))).))..))))))))).((((....((((....))))...---..)))))))-.))))................ ( -35.40) >DroVir_CAF1 15106 86 - 1 GAAGCGUUCCCAGUCGGACGAGGGCGAGAGCGAGGAGCGCGUAAAGCGCUUGACAGUCUCUUGAUAGAGAUCUAUGAG---UA-U----------------CGGA------U .......(((..(((........((....))...((((((.....))))))))).((((((....)))))).......---..-.----------------.)))------. ( -25.10) >DroSec_CAF1 16446 105 - 1 AAAGCGAUCCCAGUCGGAUGUGGGUGAGAGCGAAGAUCGCGUAAAGCGCCUGACAGUCUCUUGAUAGAGAUCUAUAGA---CAGGACGGG-UUUGUAGAUAUACAA---UGU ...((((.(((.(((...((((((((...(((.....)))......))))).)))((((((....)))))).......---...))))))-.))))..........---... ( -29.10) >DroSim_CAF1 15988 105 - 1 AAAGCGAUCCCAGUCGGAUGUGGGUGAGAGCGAAGAUCGCGUAAAGCGCCUGACAGUCUCUUGAUAGAGAUCUAUAGA---CAGGACGGG-UUUGUAGAUAUACAA---UGC ...((((.(((.(((...((((((((...(((.....)))......))))).)))((((((....)))))).......---...))))))-.))))..........---... ( -29.10) >DroWil_CAF1 19763 85 - 1 AAAGCGAUCCCAAUCGGAUGUGGGUGAGAGCGAGGAUCGCGUAAAGCGCUUUACAGUCUCUUGAUAGAGAUCUAAAU----------------UGUA--------A---UUU ...((((((((..(((....)))((....))).)))))))(((((....))))).((((((....))))))......----------------....--------.---... ( -22.30) >DroAna_CAF1 11484 107 - 1 GAAGCGGUCCCAAUCGGAUGUGGGUGAGAGCGAGGAGCGCGUGAAGCGUCUGACGGUCUCUUGAUAGAGAUCUAUAGCGUAUUUCACAGAUAUUGUAUAUAUA-----GUGU ...(((.((((..(((....)))((....))).))).)))((((((((.(((..(((((((....)))))))..))))))..)))))..(((((((....)))-----)))) ( -30.80) >consensus AAAGCGAUCCCAGUCGGAUGUGGGUGAGAGCGAAGAUCGCGUAAAGCGCCUGACAGUCUCUUGAUAGAGAUCUAUAGA___CAGGACGGG_UUUGUAGAUAUACAA___UGU ...(((((((((........)))((....))...)))))).........(((...((((((....))))))...)))................................... (-18.55 = -18.05 + -0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:41 2006