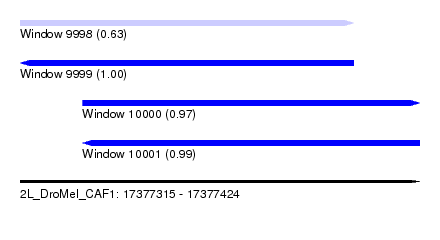
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,377,315 – 17,377,424 |
| Length | 109 |
| Max. P | 0.999912 |

| Location | 17,377,315 – 17,377,406 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 98.02 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
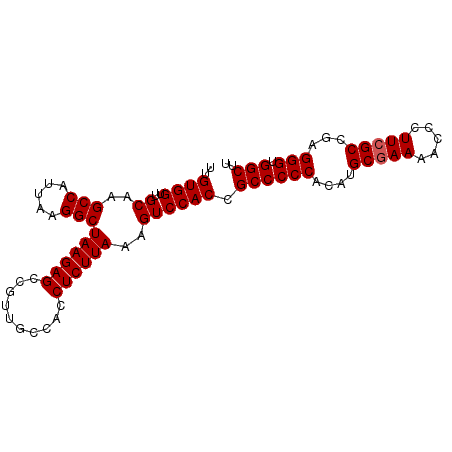
>2L_DroMel_CAF1 17377315 91 + 22407834 UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACUUGCGAAAACCCUUCGCCGAGGGUGGCUU ..((((..((..(((.....)))((((((..........))))))..)))))).((((((.(..(((((.....))))).).))).))).. ( -28.00) >DroSec_CAF1 13474 91 + 1 UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACUUGCGAAAACCCUUCGCCGAGGGUGGCUU ..((((..((..(((.....)))((((((..........))))))..)))))).((((((.(..(((((.....))))).).))).))).. ( -28.00) >DroSim_CAF1 13749 91 + 1 UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUU ..((((..((..(((.....)))((((((..........))))))..)))))).((((((.(..(((((.....))))).).))).))).. ( -28.00) >DroEre_CAF1 12996 91 + 1 UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAGGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUU ....((((...((((.....))))...))))...((((((.(((..((....))..........(((((.....))))).))))))))).. ( -30.00) >DroYak_CAF1 13413 90 + 1 UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGC-AAAACCCUUCGCAGAGGGUGGCUU .(((((..((..(((.....)))((((((..........)))))).........))..))))).((-...((((((....)))))).)).. ( -26.70) >consensus UUGUGGUUGCAAGCCAUUAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUU ..((((..((..(((.....)))((((((..........))))))..)))))).((((((....(((((.....)))))...))).))).. (-26.70 = -26.90 + 0.20)

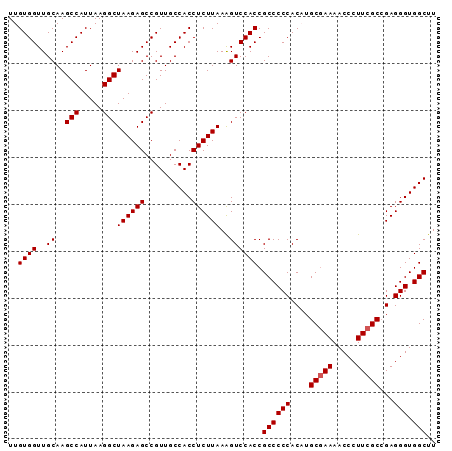
| Location | 17,377,315 – 17,377,406 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 98.02 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -37.72 |
| Energy contribution | -37.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
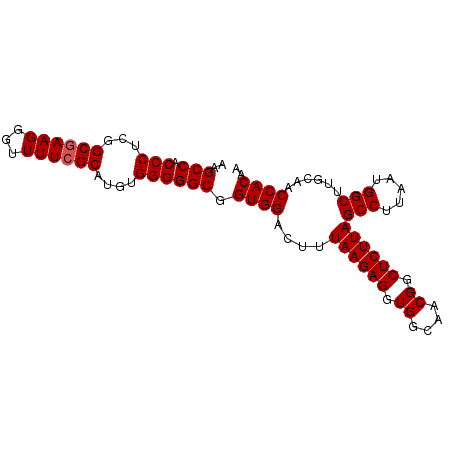
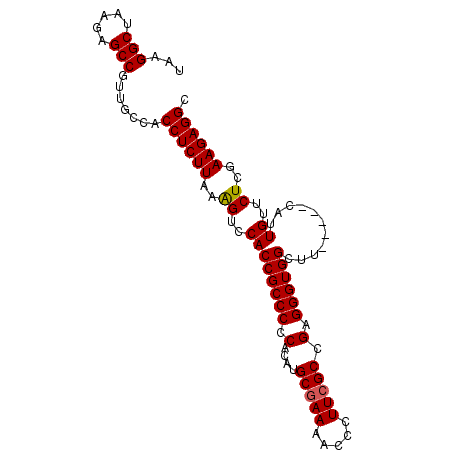
>2L_DroMel_CAF1 17377315 91 - 22407834 AAGCCACCCUCGGCGAAGGGUUUUCGCAAGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((...((((((...))))))....)))))).((((....((((((.((....)).))))))(((.....)))......)))).. ( -39.10) >DroSec_CAF1 13474 91 - 1 AAGCCACCCUCGGCGAAGGGUUUUCGCAAGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((...((((((...))))))....)))))).((((....((((((.((....)).))))))(((.....)))......)))).. ( -39.10) >DroSim_CAF1 13749 91 - 1 AAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((.((((((((...)))))).)).)))))).((((....((((((.((....)).))))))(((.....)))......)))).. ( -40.00) >DroEre_CAF1 12996 91 - 1 AAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACCUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((.((((((((...)))))).)).)))))).((((....((((((.((....)).))))))(((.....)))......)))).. ( -40.00) >DroYak_CAF1 13413 90 - 1 AAGCCACCCUCUGCGAAGGGUUUU-GCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((.(((((((....)))-))).).)))))).((((....((((((.((....)).))))))(((.....)))......)))).. ( -38.40) >consensus AAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUAAUGGCUUGCAACCACAA ..(((.(((...((((((...))))))....)))))).((((....((((((.((....)).))))))(((.....)))......)))).. (-37.72 = -37.92 + 0.20)

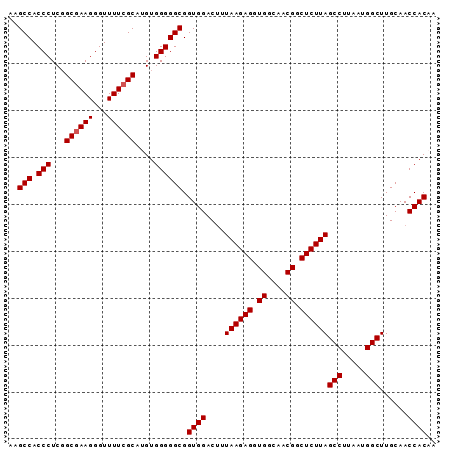
| Location | 17,377,332 – 17,377,424 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17377332 92 + 22407834 UAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACUUGCGAAAACCCUUCGCCGAGGGUGGCUU-----CAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((..((..((((((.(..(((((.....))))).).))).)))..-----...))..))..)))))). ( -29.80) >DroSec_CAF1 13491 92 + 1 UAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACUUGCGAAAACCCUUCGCCGAGGGUGGCUU-----CAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((..((..((((((.(..(((((.....))))).).))).)))..-----...))..))..)))))). ( -29.80) >DroSim_CAF1 13766 92 + 1 UAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUU-----CAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((..((..((((((.(..(((((.....))))).).))).)))..-----...))..))..)))))). ( -29.80) >DroEre_CAF1 13013 97 + 1 UAAGGCUAAGAGCCGUUGCCACCUCUUAAGGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUUCACUUCAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((....))((((((.(..(((((.....))))).).))).)))..................)))))). ( -29.60) >DroYak_CAF1 13430 91 + 1 UAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGC-AAAACCCUUCGCAGAGGGUGGCUU-----CAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((..((..((((((...(((-..........)))..))).)))..-----...))..))..)))))). ( -27.00) >consensus UAAGGCUAAGAGCCGUUGCCACCUCUUAAAGUCCACCGCCCCCACAUGCGAAAACCCUUCGCCGAGGGUGGCUU_____CAUUGUUCUCGAAGAGGC ...(((.....))).......((((((..((..((((((((.(....(((((.....))))).).))))))...........))..))..)))))). (-27.70 = -27.74 + 0.04)

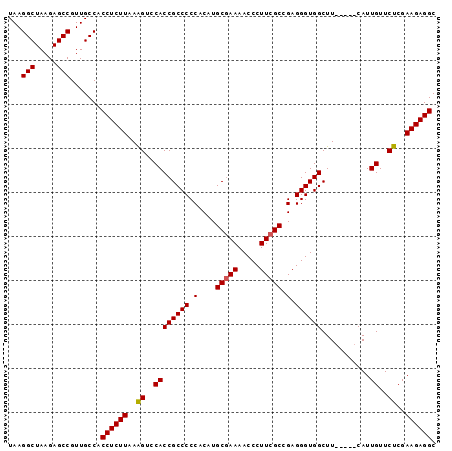
| Location | 17,377,332 – 17,377,424 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -32.04 |
| Energy contribution | -32.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
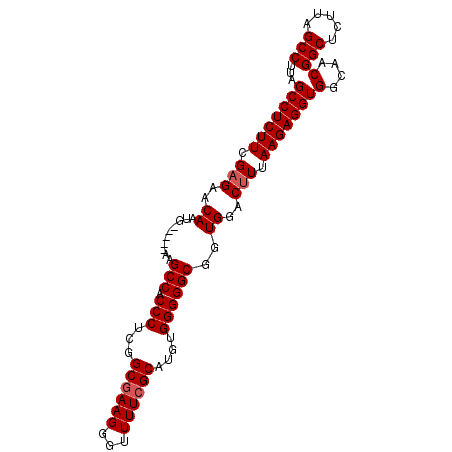
>2L_DroMel_CAF1 17377332 92 - 22407834 GCCUCUUCGAGAACAAUG-----AAGCCACCCUCGGCGAAGGGUUUUCGCAAGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((((((.(((..((...-----..(((.(((...((((((...))))))....))))))..))..))).)))))))(....)(((.....)))... ( -36.30) >DroSec_CAF1 13491 92 - 1 GCCUCUUCGAGAACAAUG-----AAGCCACCCUCGGCGAAGGGUUUUCGCAAGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((((((.(((..((...-----..(((.(((...((((((...))))))....))))))..))..))).)))))))(....)(((.....)))... ( -36.30) >DroSim_CAF1 13766 92 - 1 GCCUCUUCGAGAACAAUG-----AAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((((((.(((..((...-----..(((.(((.((((((((...)))))).)).))))))..))..))).)))))))(....)(((.....)))... ( -37.20) >DroEre_CAF1 13013 97 - 1 GCCUCUUCGAGAACAAUGAAGUGAAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACCUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((.((((.................(((.(((.((((((((...)))))).)).))))))((....))....)))).)))...(((.....)))... ( -34.70) >DroYak_CAF1 13430 91 - 1 GCCUCUUCGAGAACAAUG-----AAGCCACCCUCUGCGAAGGGUUUU-GCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((((((.(((..((...-----..(((.(((.(((((((....)))-))).).))))))..))..))).)))))))(....)(((.....)))... ( -35.60) >consensus GCCUCUUCGAGAACAAUG_____AAGCCACCCUCGGCGAAGGGUUUUCGCAUGUGGGGGCGGUGGACUUUAAGAGGUGGCAACGGCUCUUAGCCUUA (((((((.(((..((..........(((.(((...((((((...))))))....))))))..))..))).)))))))(....)(((.....)))... (-32.04 = -32.44 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:35 2006