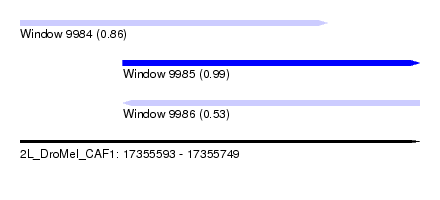
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,355,593 – 17,355,749 |
| Length | 156 |
| Max. P | 0.988365 |

| Location | 17,355,593 – 17,355,713 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17355593 120 + 22407834 UUCCCCUCAAAACCUUCCUCAGUUUGUCUGUUUGGUGAAGCAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGACUAACAGCUUCAAACUCUCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... ( -22.40) >DroSec_CAF1 57437 120 + 1 UUUCCCUCAAAACCUUCCUCAGUUUGUCUGUUUGGUGAAGCAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... ( -21.80) >DroSim_CAF1 58034 120 + 1 UUCCCCUCAAAACCUUCCUCAGUUUGUCUGUUUGGUGAAGCAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... ( -21.80) >DroEre_CAF1 55194 120 + 1 UUCCCCUCAAAACCUUCCUCAGUUUGUCUGUUUGGUGAAGCGAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCCCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... ( -21.50) >DroYak_CAF1 51585 120 + 1 UUCCCCUUAAAACCUUCCACAGUUUGUCUGUUUGGUGAAGCGAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... ( -21.50) >DroAna_CAF1 58030 114 + 1 CUCCCCUC------UCUCUCUCCCUGUCUGUUUGGUGAAGCAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCAACUCC ........------..........((...((((((..(((.((((....)))))))..)(((((.(((((...........)))))((.....))....))))).)))))...))..... ( -19.40) >consensus UUCCCCUCAAAACCUUCCUCAGUUUGUCUGUUUGGUGAAGCAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCC .....................(((.(((((...(((...((((...((((......(((((....)))))....))))..)))).)))...))))).))).................... (-20.74 = -20.88 + 0.14)


| Location | 17,355,633 – 17,355,749 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17355633 116 + 22407834 CAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGACUAACAGCUUCAAACUCUCGACUCCUUUGGCCCAGGAAUUUUCAGCUACACAACACAACA--C-- ................((.(((((((((((...........)))))((((...(.((((..(..((........))..)..))))).)))).....)))))).))..........--.-- ( -17.40) >DroSec_CAF1 57477 116 + 1 CAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCCUUUGGCCCAGGAAUUUUCAGCUACACAACACAACA--C-- ................((.(((((((((((...........)))))((((...((((((..(..((........))..)..)))))))))).....)))))).))..........--.-- ( -24.00) >DroSim_CAF1 58074 116 + 1 CAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCCUUUGGCCCAGGAAUUUUCAGCUACACAACACAACA--C-- ................((.(((((((((((...........)))))((((...((((((..(..((........))..)..)))))))))).....)))))).))..........--.-- ( -24.00) >DroEre_CAF1 55234 116 + 1 CGAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCCCGACUCCUUUGGCCCAGGAAUUUUCAGCUACACAACGCAACA--C-- .((((.....)))).(((((((((((((((...........)))))((((...((((((..(..((........))..)..)))))))))).....)))))).......))))..--.-- ( -24.61) >DroYak_CAF1 51625 111 + 1 CGAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCCUUUGGCCCAAGAAUUUUCAGCUACACA-----ACA--C-- .((((.....))))..((.(((((((((((...........))))).......((((((..(..((........))..)..)))))).........)))))).))..-----...--.-- ( -19.40) >DroAna_CAF1 58064 120 + 1 CAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCAACUCCUUUGGCCCAGGAAUUUUCAGCAACACACCGAGCCAUUCCC ....................((((((((((...........)))))((((...((((((......................)))))))))).....)))))................... ( -20.35) >consensus CAAAACCUAUUUUCUUUGCAGCUGAUUGCAUUUUAUAGCAUUGCAACCUGUCAGGCUAACAGCUUCAAACUCUCGACUCCUUUGGCCCAGGAAUUUUCAGCUACACAACACAACA__C__ ................((.(((((((((((...........)))))((((...((((((..(..((........))..)..)))))))))).....)))))).))............... (-20.32 = -20.98 + 0.67)


| Location | 17,355,633 – 17,355,749 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -26.91 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
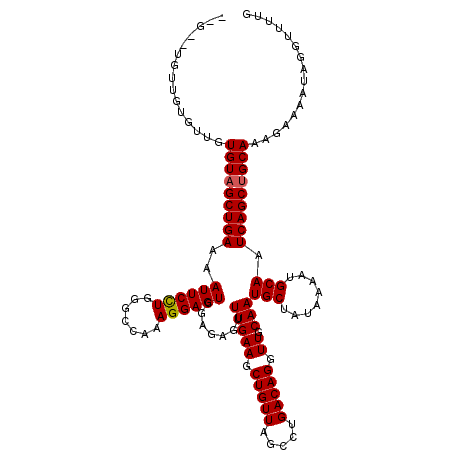
>2L_DroMel_CAF1 17355633 116 - 22407834 --G--UGUUGUGUUGUGUAGCUGAAAAUUCCUGGGCCAAAGGAGUCGAGAGUUUGAAGCUGUUAGUCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUUG --.--((((......(((((((((..((((((.......)))))).......(((((.(((((.....))))).)).)))(((........))).))))))))).....))))....... ( -28.90) >DroSec_CAF1 57477 116 - 1 --G--UGUUGUGUUGUGUAGCUGAAAAUUCCUGGGCCAAAGGAGUCGAGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUUG --.--((((......(((((((((.....(((((((.((.((..((((....))))..)).)).)))...))))(((((.(.......).)))))))))))))).....))))....... ( -29.50) >DroSim_CAF1 58074 116 - 1 --G--UGUUGUGUUGUGUAGCUGAAAAUUCCUGGGCCAAAGGAGUCGAGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUUG --.--((((......(((((((((.....(((((((.((.((..((((....))))..)).)).)))...))))(((((.(.......).)))))))))))))).....))))....... ( -29.50) >DroEre_CAF1 55234 116 - 1 --G--UGUUGCGUUGUGUAGCUGAAAAUUCCUGGGCCAAAGGAGUCGGGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUCG --.--(((.(((((((.(((((..((((((((((.((...))..))))))))))..)))))..((((.....))))))))))).)))...((((......))))..((((.....)))). ( -35.00) >DroYak_CAF1 51625 111 - 1 --G--UGU-----UGUGUAGCUGAAAAUUCUUGGGCCAAAGGAGUCGAGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUCG --.--...-----..(((((((((((((((((((.((...))..)))))))))).((.(((((.....))))).))....(((........))).)))))))))..((((.....)))). ( -31.20) >DroAna_CAF1 58064 120 - 1 GGGAAUGGCUCGGUGUGUUGCUGAAAAUUCCUGGGCCAAAGGAGUUGAGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUUG .....((((((((.(.(((......))).)))))))))..(.((((((..((((..((((((.((((.....)))))))..)))...))))....)))))).)................. ( -30.70) >consensus __G__UGUUGUGUUGUGUAGCUGAAAAUUCCUGGGCCAAAGGAGUCGAGAGUUUGAAGCUGUUAGCCUGACAGGUUGCAAUGCUAUAAAAUGCAAUCAGCUGCAAAGAAAAUAGGUUUUG ...............(((((((((..((((((.......)))))).......(((((.(((((.....))))).)).)))(((........))).)))))))))................ (-26.91 = -26.93 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:21 2006