
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,354,179 – 17,354,306 |
| Length | 127 |
| Max. P | 0.956040 |

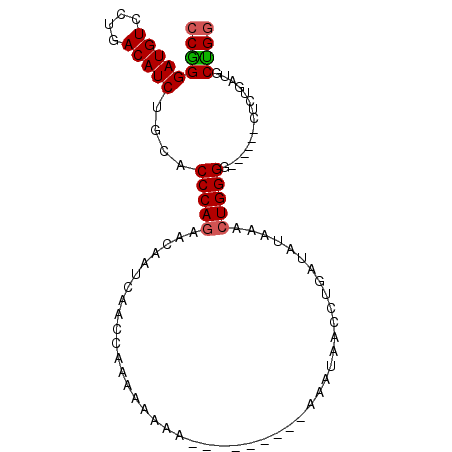
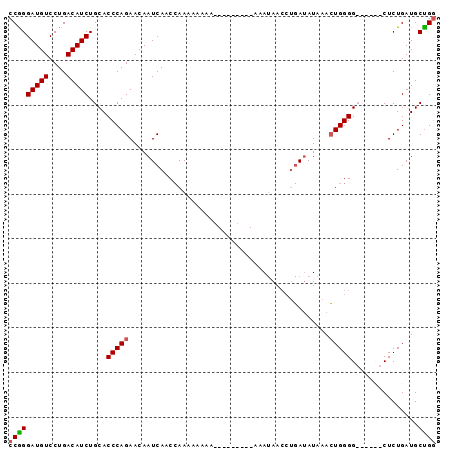
| Location | 17,354,179 – 17,354,275 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -17.14 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17354179 96 - 22407834 CCAGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAACAAAAAAAAAUAAAUAAAUAACCUGAUAUAAACUGGGGCCCUCGCUCUGAUGCUGG (((((((((....))))).((.(((((....((((............................)))).....))))))).............)))) ( -18.39) >DroSec_CAF1 56058 81 - 1 CCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAAAAA---------AAAUAACCUGAUAUAAACUGGGG------CUCUGAUGCUGG .((((....)))).((((.((.(((((....((((...........---------........)))).....))))))------)...)))).... ( -18.91) >DroSim_CAF1 52412 80 - 1 CCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAA-AA---------AAAUAACCAGAUAUAAACUGGGG------CUCUGAUGCUGG .((((....)))).((((.((.(((((....(((.........-..---------.........))).....))))))------)...)))).... ( -17.75) >DroEre_CAF1 53849 85 - 1 CCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACGAAAAGAAA---------AAAAAA--UUAAAUAAAAUGGGGCCCUCACUCUGAUGCCGC .((((((((....))))).((.((((....................---------......--..........)))))).............))). ( -13.52) >consensus CCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAAAAA_________AAAUAACCUGAUAUAAACUGGGG______CUCUGAUGCUGG (((((((((....)))))....(((((.............................................)))))...............)))) (-12.73 = -12.86 + 0.13)

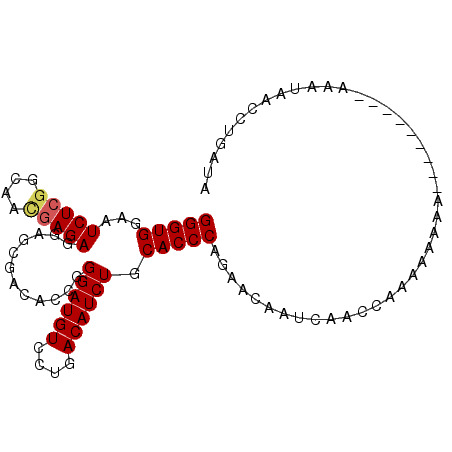
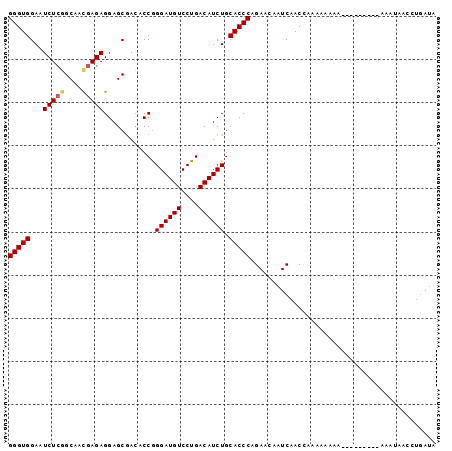
| Location | 17,354,207 – 17,354,306 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17354207 99 - 22407834 GGGUGGAAUCUCUGCAACGAGAGGAGCAAAACCAGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAACAAAAAAAAAUAAAUAAAUAACCUGAUA (((((.......(((..(....)..)))......((((((....)))))).)))))........................................... ( -20.30) >DroSec_CAF1 56080 90 - 1 GGGUGGAAUCUAGGCAACGAGAGGAGCGACACCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAAAAA---------AAAUAACCUGAUA (((((........((..(....)..)).......((((((....)))))).))))).....................---------............. ( -19.60) >DroSim_CAF1 52434 89 - 1 GGGUGGAAUCUCGGCAAUGAGAGGAGCGACACCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAA-AA---------AAAUAACCAGAUA (((((...(((((....)))))............((((((....)))))).)))))..................-..---------............. ( -19.80) >DroEre_CAF1 53877 88 - 1 GGGUGGAAUCUCGGCUUCGAGAGGAGCGACACCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACGAAAAGAAA---------AAAAAA--UUAAA (((((........(((((....))))).......((((((....)))))).))))).....................---------......--..... ( -24.20) >consensus GGGUGGAAUCUCGGCAACGAGAGGAGCGACACCGGGAUGUCCUGACAUCUGCACCCAGAACAAUCAACCAAAAAAAA_________AAAUAACCUGAUA (((((...(((((....)))))............((((((....)))))).)))))........................................... (-19.20 = -19.57 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:18 2006