| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,341,530 – 17,341,632 |
| Length | 102 |
| Max. P | 0.878159 |

| Location | 17,341,530 – 17,341,632 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.50 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.10 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17341530 102 + 22407834 ACUCAGCG---CCAGCUGAGGUUGGCCAACAGGU-CAGCAGAGUGUUUAGGGUGAAUGCGGAAUUGGUAACAG--UUU---AGG----UUGUAUUCAGAGCAUCUC-----AUGAUAUUU .((((((.---...))))))(((((((....)))-)))).((((((((.(((((...((.((((((....)))--)))---..)----)..))))).))))).)))-----......... ( -37.30) >DroSec_CAF1 37884 102 + 1 ACUCAGCG---CCAGCUGAGGUUGGCCAACAGGU-CAGCAGAGUGUUUAGGGUGAAUGCGAAAUUGGUAACAG--CUC---AGG----UUAUAUACAGUUCAUCUC-----AUGAUAUCU .((((((.---...))))))(((((((....)))-))))(((.((((..((((((((.........(((((..--...---..)----)))).....)))))))).-----..))))))) ( -31.94) >DroSim_CAF1 40523 102 + 1 ACUCAGCG---CCAGCUGAGGUUGGCCAACAGGU-CAGCAGAGUGUUUAGGGUGAAUGCGGAAUUGGUAACAG--CUC---AGG----UUAUAUUCAGAGCAUCUC-----GUGAUAUCU .((((((.---...))))))(((((((....)))-)))).((((((((.(((((...((.((.(((....)))--.))---..)----)..))))).))))).)))-----......... ( -34.10) >DroEre_CAF1 40061 93 + 1 ACUCAGCG---CAA-CUGAAGUUGGCCAACAGGUUUAGCAGGGUGUUUAGGGUUAAUGCGGAGUUGGUAACAA--UGC---CAGCGCAUUAUAUCAAAAU-----------AA------- .((.((((---(..-(((((.(((.....))).)))))....))))).))(((((((((...(((((((....--)))---)))))))))).))).....-----------..------- ( -26.50) >DroYak_CAF1 34911 96 + 1 ACUCAGCG---CAAGCUGAGGUUGGCCAACGGGUUCAGCAGGGUGUAUUGGGUUAAUACACACUUGGUAACAG--UUU---UAG----UUAUACUUAGAU-----------AUUAUACU- .((((((.---...))))))(((((((....)).)))))(((((((((((...)))))))).))).(((((..--...---..)----))))........-----------........- ( -29.00) >DroAna_CAF1 46644 105 + 1 -CCCGGUGGGUGCAGCUGAGCCUGGCUACCAGGAAGAGUUGGGUAUUUUGGGACUACCCAAGAUUGGUAACACAGAUCUGUAGA----UUAGA---AUUUUAUCUGGAAAUAA------- -.((((...((((((((...(((((...)))))...)))))...((((((((....))))))))......)))...((((....----.))))---.......))))......------- ( -28.40) >consensus ACUCAGCG___CCAGCUGAGGUUGGCCAACAGGU_CAGCAGAGUGUUUAGGGUGAAUGCGGAAUUGGUAACAG__UUC___AGG____UUAUAUUCAGAUCAUCUC_____AUGAUAU__ .((((((.......))))))(((((((....))).))))................................................................................. (-10.73 = -11.10 + 0.37)

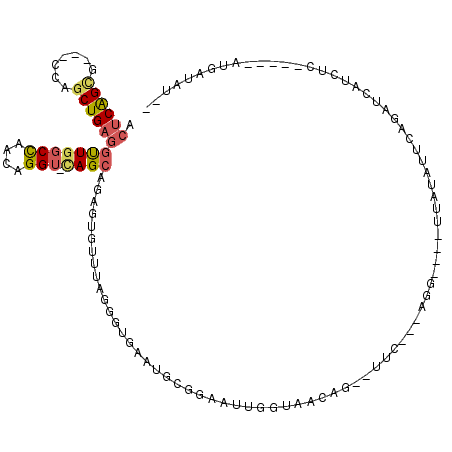

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:06 2006