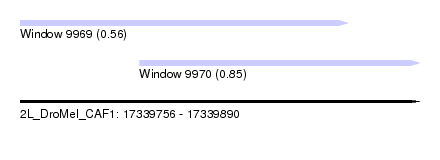
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,339,756 – 17,339,890 |
| Length | 134 |
| Max. P | 0.849945 |

| Location | 17,339,756 – 17,339,866 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -25.40 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
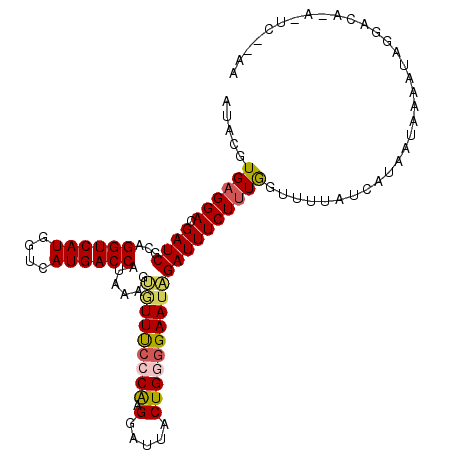
>2L_DroMel_CAF1 17339756 110 + 22407834 AUGUCCCUAGUGGCCACAUCCACGUCCCGUUUUUCGACGAAUACGUGAGGACGA-U------CGCAGGUCAUGGUCA-UGACCAUAAAGUGUUUCCCAA-GGAUUACUGGGGAAUAGA-U ...((((((((((((.......((((.........))))(((((((((......-)------))).((((((....)-))))).....)))))......-)).)))))))))).....-. ( -36.20) >DroSec_CAF1 36078 110 + 1 AUGUCCCUAGUGGCCAUAUCCACGUCCCGUUUUUCGACGGAUACGUGAGGACGA-U------CGCAGGUCAUGGUCA-UGACCAUAAAGUGUUUCCCAA-GGAUUACUGGGGAAUAGA-U ...((((((((((((.....(((((.(((((....)))))..))))).(((...-.------(((.((((((....)-))))).....)))..)))...-)).)))))))))).....-. ( -39.00) >DroSim_CAF1 38705 110 + 1 AUGUCCCUAGUGGCCAUAUCCACGUCCCGUUUUUCGACGGAUACGUGAGGACGA-U------CGCAGGUCAUGGUCA-UGACCAUAAAGUGUUUCCCAA-GGAUUACUGGGGAAUAGA-U ...((((((((((((.....(((((.(((((....)))))..))))).(((...-.------(((.((((((....)-))))).....)))..)))...-)).)))))))))).....-. ( -39.00) >DroEre_CAF1 38205 109 + 1 AUGUCCCUAGUGGCCAUAUCCACGUCCCGUUUUUCGACGGAUACGUGAGGACGA-U------CGCAGGUCAUGGUCA-UGACCAUAAGGGAUUCCACAC-GGAUUACUGG-GAAUAGA-U .(((.((((((((((.....(((((.(((((....)))))..))))).(((..(-(------(.(.((((((....)-)))))....).))))))....-)).)))))))-).)))..-. ( -36.90) >DroYak_CAF1 33146 110 + 1 AUGUCCCUAGUGGCCAUAUCCACGUCCCGUUUUUCGACGGAUACGUGAGGAGGA-U------CGCAGGUCAUGGUCA-UGACCAUAAGGGAUUCCACGC-GGAUUACUGGGGAACCGA-U ...((((((((((((.....(((((.(((((....)))))..))))).((((..-(------(...((((((....)-)))))....))..))))....-)).)))))))))).....-. ( -43.30) >DroAna_CAF1 43987 117 + 1 GUGUCCCUGUUGGCCAU--GCACGUUCCGUUUUUCGCCGCGUACGUGAGGAUGGGUUGCUCCUGAAGGCCAUGGGCAGUGCCUAGAAAGGGUGUUCUAGCAGAGAGCUUU-AAAAAUAUU ....((((..(((((..--.(((((..(((........))).)))))((((........))))...)))))(((((...)))))...)))).(((((.....)))))...-......... ( -33.70) >consensus AUGUCCCUAGUGGCCAUAUCCACGUCCCGUUUUUCGACGGAUACGUGAGGACGA_U______CGCAGGUCAUGGUCA_UGACCAUAAAGGGUUUCCCAA_GGAUUACUGGGGAAUAGA_U ...((((((((((((.....(((((.(((((....)))))..)))))....((.........))..))))((((((...))))))....................))))))))....... (-25.40 = -26.15 + 0.75)


| Location | 17,339,796 – 17,339,890 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17339796 94 + 22407834 AUACGUGAGGACGAUCGCAGGUCAUGGUCAUGACCAUAAAGUGUUUCCCAAGGAUUACUGGGGAAUAGAUUUCUUUGGUUUUAUCAUAAUAAAA---------------- ......(((((.((((...((((((....))))))......(((((((((.(.....)))))))))))))))))))..................---------------- ( -24.70) >DroSec_CAF1 36118 100 + 1 AUACGUGAGGACGAUCGCAGGUCAUGGUCAUGACCAUAAAGUGUUUCCCAAGGAUUACUGGGGAAUAGAUUUCUUUGGUUUUGUCAU---AAACAAGGAUU-------AA ....((((......)))).((((((....))))))......(((((((((.(.....))))))))))....((((((..........---...))))))..-------.. ( -27.02) >DroSim_CAF1 38745 110 + 1 AUACGUGAGGACGAUCGCAGGUCAUGGUCAUGACCAUAAAGUGUUUCCCAAGGAUUACUGGGGAAUAGAUUUCUUUGGUUUUAUCAUAAUAAACUAGGACAAAUUCGCAA ....(((((((.((((...((((((....))))))......(((((((((.(.....))))))))))))))))((((((((.........)))))))).....))))).. ( -29.40) >DroEre_CAF1 38245 109 + 1 AUACGUGAGGACGAUCGCAGGUCAUGGUCAUGACCAUAAGGGAUUCCACACGGAUUACUGG-GAAUAGAUUUCUUUAGUUCUAUCAUUAUAAAUUGGGACAUAUUCCUAA ...((((.(((..(((.(.((((((....))))))....).)))))).))))......(((-((((((....)....((((((...........)))))).)))))))). ( -29.50) >DroYak_CAF1 33186 110 + 1 AUACGUGAGGAGGAUCGCAGGUCAUGGUCAUGACCAUAAGGGAUUCCACGCGGAUUACUGGGGAACCGAUUUCUCUAGCUUUAUCAUUAUAAAAUGGGAUACAAUCACAA ...((((.((((..((...((((((....))))))....))..)))).))))((((.((((((((.....)))))))).....(((((....))))).....)))).... ( -31.00) >consensus AUACGUGAGGACGAUCGCAGGUCAUGGUCAUGACCAUAAAGUGUUUCCCAAGGAUUACUGGGGAAUAGAUUUCUUUGGUUUUAUCAUAAUAAAAUAGGACA_A_UC__AA .....((((((.((((...((((((....))))))......(((((((((.(.....))))))))))))))))))))................................. (-21.48 = -21.68 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:05 2006