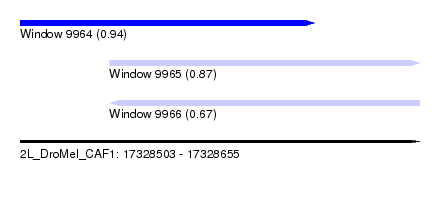
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,328,503 – 17,328,655 |
| Length | 152 |
| Max. P | 0.935345 |

| Location | 17,328,503 – 17,328,615 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -33.15 |
| Energy contribution | -33.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
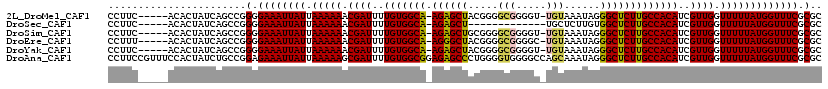
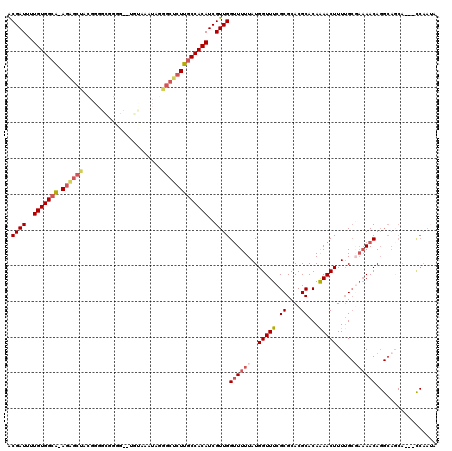
>2L_DroMel_CAF1 17328503 112 + 22407834 CCUUC-----ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA-AGAGCUACGGGGCGGGGU-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .....-----.............(.((((((((.(((((.((((..(((((((-((((((.....(((....-)))......)))))))))))))..)))).))))))))))))).).. ( -38.20) >DroSec_CAF1 26006 100 + 1 CCUUC-----ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA-AGAGCU-------------UGCUCUUGUGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .....-----.............(.((((((((.(((((.((((..(((((((-((((((-------------.((....)))))))))))))))..)))).))))))))))))).).. ( -37.00) >DroSim_CAF1 28970 112 + 1 CCUUC-----ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA-AGAGCUGCGGGGCGGGGU-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .....-----.............(.((((((((.(((((.((((..(((((((-((((((.....(((....-)))......)))))))))))))..)))).))))))))))))).).. ( -37.10) >DroEre_CAF1 28116 112 + 1 CCUUU-----ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA-AGGGCUACGGGGCGGGGC-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .....-----.............(.((((((((.(((((.((((..(((((((-((((((.....(((....-)))......)))))))))))))..)))).))))))))))))).).. ( -38.40) >DroYak_CAF1 20946 112 + 1 CCUUC-----ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA-AGAGCUACGGGGCGGGGU-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .....-----.............(.((((((((.(((((.((((..(((((((-((((((.....(((....-)))......)))))))))))))..)))).))))))))))))).).. ( -38.20) >DroAna_CAF1 33575 119 + 1 CCUUCCGUUUCCACUAUCUGCCGGAGAAAUUAUUAAAAAGCGAUUUUGUGGCGGAGAGCCCUGGGGUGGGGCCAGCAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC ...((((..............))))((((((((.(((((.((((..(((((((.(((((((((.(.((....)).)...))))))))))))))))..)))).))))))))))))).... ( -48.24) >consensus CCUUC_____ACACUAUCAGCCGGGGAAAUUAUUAAAAAACGAUUUUGUGGCA_AGAGCUACGGGGCGGGGU_UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGC .......................(.((((((((.(((((.((((..(((((((.((((((.....(((.....)))......)))))))))))))..)))).))))))))))))).).. (-33.15 = -33.18 + 0.03)

| Location | 17,328,537 – 17,328,655 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -36.39 |
| Consensus MFE | -22.71 |
| Energy contribution | -23.93 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
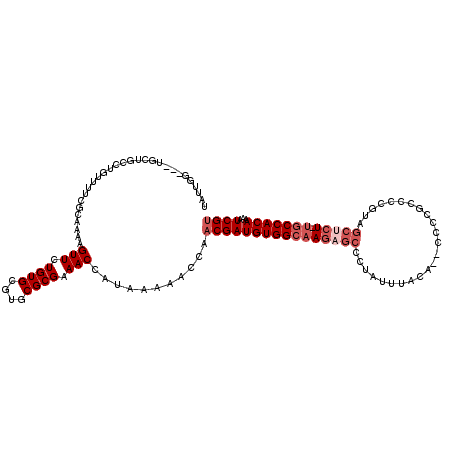
>2L_DroMel_CAF1 17328537 118 + 22407834 ACGAUUUUGUGGCA-AGAGCUACGGGGCGGGGU-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAGAACUUUUGCGAAAACAGGCAGCACCACCAAUC ((((...(((((((-((((((.....(((....-)))......)))))))))))))))))((((.....((.(((((((...............))))))).))((.....))))))... ( -36.96) >DroVir_CAF1 41706 99 + 1 ACGAUUUUGUGGGA-AC------------------GUCAGAAAGGCUUUUGCCACAUCGUUGGGUUUUAUGGUUUCGCGCACGCACAAAACUUUGGCC--AACAGGCAACAGAAUCAAAA ..(((((((((...-.)------------------(((.....(((....))).....(((((.(.....(((((.((....))...)))))..).))--))).))).)))))))).... ( -27.10) >DroSec_CAF1 26040 106 + 1 ACGAUUUUGUGGCA-AGAGCU-------------UGCUCUUGUGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAGAACUUUUGCGAAAACAGGCAGCACCACCAAUC ((((...(((((((-((((((-------------.((....)))))))))))))))))))((((.....((.(((((((...............))))))).))((.....))))))... ( -35.76) >DroEre_CAF1 28150 115 + 1 ACGAUUUUGUGGCA-AGGGCUACGGGGCGGGGC-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAGAACUUUUGCGAAAACAGGCAACA---CCAAUA .......(((((((-((((((.....(((....-)))......)))))))))))))..((((((.....((.(((((((...............))))))).))(....))---))))). ( -37.86) >DroYak_CAF1 20980 115 + 1 ACGAUUUUGUGGCA-AGAGCUACGGGGCGGGGU-UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAAAACUUUUGCGAAAACAGGCAGCA---CCAAUA .......(((((((-((((((.....(((....-)))......)))))))))))))..((((((.....((.(((((((...............))))))).))......)---))))). ( -37.16) >DroAna_CAF1 33614 102 + 1 GCGAUUUUGUGGCGGAGAGCCCUGGGGUGGGGCCAGCAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAAAACUUUUG--AAAACA---------------- .((((..(((((((.(((((((((.(.((....)).)...))))))))))))))))..))))(((((((.(((((.((....))...)))))..))--))))).---------------- ( -43.50) >consensus ACGAUUUUGUGGCA_AGAGCUACGGGGCGGGG__UGUAAAUAGGGCUCUUGCCACAUCGUUGGUUUUUAUGGUUUCGCGCACGCACAAAACUUUUGCGAAAACAGGCAGCA___CCAAUA .((((..(((((((.((((((......................)))))))))))))..))))((((((..(((((.((....))...))))).....))))))................. (-22.71 = -23.93 + 1.22)

| Location | 17,328,537 – 17,328,655 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -32.59 |
| Consensus MFE | -17.53 |
| Energy contribution | -18.86 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
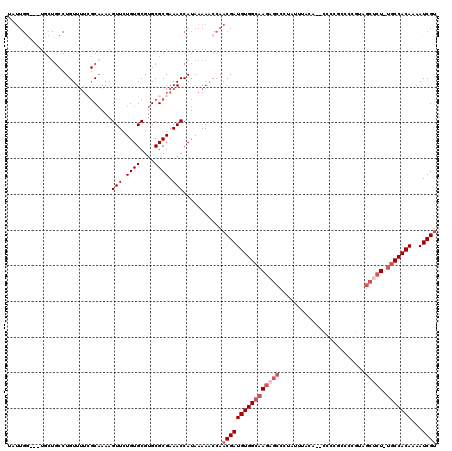
>2L_DroMel_CAF1 17328537 118 - 22407834 GAUUGGUGGUGCUGCCUGUUUUCGCAAAAGUUCUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCCUAUUUACA-ACCCCGCCCCGUAGCUCU-UGCCACAAAAUCGU ...((((((.....)).(((((((((...((.....)).)))).))))).......))))((((((((((((((((..........-.............)))))-)))))))...)))) ( -35.60) >DroVir_CAF1 41706 99 - 1 UUUUGAUUCUGUUGCCUGUU--GGCCAAAGUUUUGUGCGUGCGCGAAACCAUAAAACCCAACGAUGUGGCAAAAGCCUUUCUGAC------------------GU-UCCCACAAAAUCGU ....((((.(((.....(((--((.....((((((((....))))))))........)))))((...(((....)))..))....------------------..-....))).)))).. ( -22.22) >DroSec_CAF1 26040 106 - 1 GAUUGGUGGUGCUGCCUGUUUUCGCAAAAGUUCUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCACAAGAGCA-------------AGCUCU-UGCCACAAAAUCGU ...((((((.....)).(((((((((...((.....)).)))).))))).......))))((((((((((((((((..........-------------.)))))-)))))))...)))) ( -36.40) >DroEre_CAF1 28150 115 - 1 UAUUGG---UGUUGCCUGUUUUCGCAAAAGUUCUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCCUAUUUACA-GCCCCGCCCCGUAGCCCU-UGCCACAAAAUCGU ...(((---(.......(((((((((...((.....)).)))).))))).......))))(((((((((((((.((..........-.............)).))-)))))))...)))) ( -30.54) >DroYak_CAF1 20980 115 - 1 UAUUGG---UGCUGCCUGUUUUCGCAAAAGUUUUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCCUAUUUACA-ACCCCGCCCCGUAGCUCU-UGCCACAAAAUCGU ...(((---(..(((........)))...((((((((....)))))))).......))))((((((((((((((((..........-.............)))))-)))))))...)))) ( -35.90) >DroAna_CAF1 33614 102 - 1 ----------------UGUUUU--CAAAAGUUUUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCCUAUUUGCUGGCCCCACCCCAGGGCUCUCCGCCACAAAAUCGC ----------------.(((((--.....((((((((....))))))))....)))))...(((((((((.((((((((....(.((....)).)..))))))))..))))))...))). ( -34.90) >consensus UAUUGG___UGCUGCCUGUUUUCGCAAAAGUUCUGUGCGUGCGCGAAACCAUAAAAACCAACGAUGUGGCAAGAGCCCUAUUUACA__CCCCGCCCCGUAGCUCU_UGCCACAAAAUCGU .............................(((.((((....)))).)))...........((((((((((((((((........................))))).)))))))...)))) (-17.53 = -18.86 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:01 2006