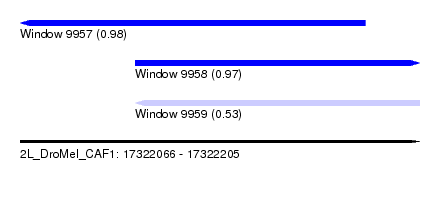
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,322,066 – 17,322,205 |
| Length | 139 |
| Max. P | 0.977720 |

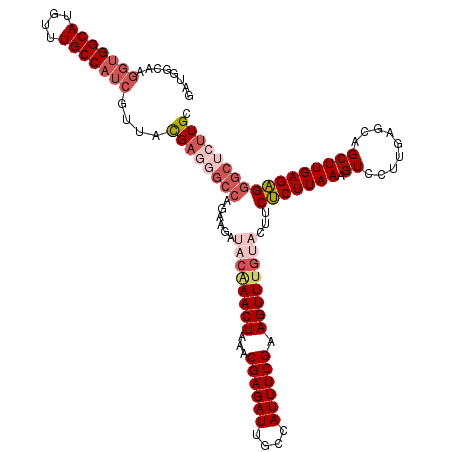
| Location | 17,322,066 – 17,322,186 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -29.98 |
| Energy contribution | -32.78 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
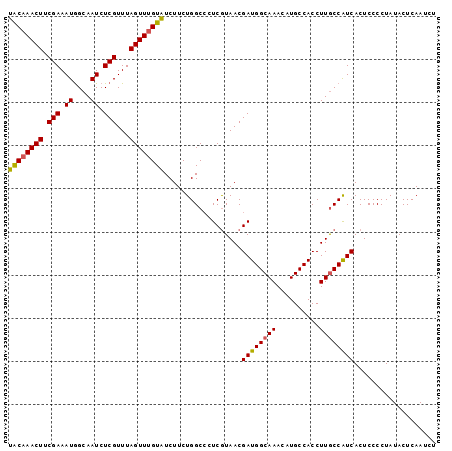
>2L_DroMel_CAF1 17322066 120 - 22407834 GAUGGCAAGGUGGCAUGUUUGCCAUCGUUAUGAGGGCCAGAAGAUACGAACUAAACGAGAUUGCCAUUUCGAAGUUUGUACUUCUCUUAAAGUCCUUGAGCAGCUUGAGAGGGCUCUUGC ....(((((((((((....)))))))(((.(((((((.(((((.((((((((...((((((....)))))).)))))))))))))......))))))))))(((((....))))).)))) ( -42.50) >DroSec_CAF1 19723 118 - 1 GACGGCAAGGUGGCAUGUUUGCCAUCGUUACGA--GCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUACUUCUCUUAAAGUCCUUGAGCAGCUUGAGAGGGCUCUUGC ....(((((((((((....))))))).....((--((((((((.((((((((...((((((....)))))).))))))))))))).........((..((...))..))..))))))))) ( -42.40) >DroSim_CAF1 22071 120 - 1 GAUGGUAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUACUUCUCUUAAAGUCCUUGAGCAGCUUGAGAGGGCUCUUGC ....(((((((((((....))))))).))))((((((.(((((.((((((((...((((((....)))))).)))))))))))))......))))))(((.(((((....)))))))).. ( -44.20) >DroEre_CAF1 21634 93 - 1 -----------GGCAUGUUUGCCAUCGUUACG----------------AACUAAACGAGAUUGCCAUUUCGAAGUUUGUACUGCGCUUAAAGUCCUUGAGCAGCUUGAGCGGGCUCUUGC -----------((((....))))...((((((----------------((((...((((((....)))))).))))))))..))((((((.....)))))).((..(((....)))..)) ( -27.90) >DroYak_CAF1 14698 120 - 1 GAUGGCAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGACACAAACUAAACGAGAUUGCUAUUUCGAAGUUUGCACUUCUCUUAAAGUCCUUAAGCAGCUUGAGAGGGCUCUUGC (((((((((........)))))))))....(((((((((((((...((((((...((((((....)))))).))))))..))))).........((((((...))))))..)))))))). ( -41.40) >consensus GAUGGCAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUACUUCUCUUAAAGUCCUUGAGCAGCUUGAGAGGGCUCUUGC ........(((((((....)))))))....((((((((......((((((((...((((((....)))))).))))))))...(((((((.((.........))))))))))))))))). (-29.98 = -32.78 + 2.80)

| Location | 17,322,106 – 17,322,205 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
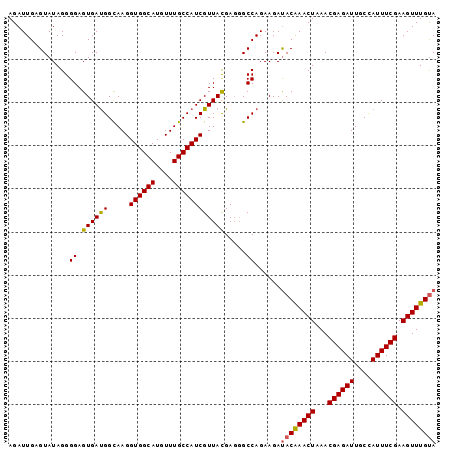
>2L_DroMel_CAF1 17322106 99 + 22407834 UACAAACUUCGAAAUGGCAAUCUCGUUUAGUUCGUAUCUUCUGGCCCUCAUAACGAUGGCAAACAUGCCACCUUGCCAUCACUCCCCUAUACUCAAUCU .............(((((((..(((((......((........))......)))))(((((....)))))..))))))).................... ( -17.20) >DroSec_CAF1 19763 97 + 1 UACAAACUUCGAAAUGGCAAUCUCGUUUAGUUUGUAUCUUCUGGC--UCGUAACGAUGGCAAACAUGCCACCUUGCCGUCACUCCCCUAUACUCAAUCU ((((((((.(((.((....)).)))...)))))))).....((((--..((((.(.(((((....)))))).)))).)))).................. ( -21.40) >DroSim_CAF1 22111 99 + 1 UACAAACUUCGAAAUGGCAAUCUCGUUUAGUUUGUAUCUUCUGGCCCUCGUAACGAUGGCAAACAUGCCACCUUACCAUCACUCCCCUAUACUCAAUCU ((((((((.(((.((....)).)))...)))))))).............((((.(.(((((....)))))).))))....................... ( -18.20) >DroYak_CAF1 14738 99 + 1 UGCAAACUUCGAAAUAGCAAUCUCGUUUAGUUUGUGUCUUCUGGCCCUCGUAACGAUGGCAAACAUGCCACCUUGCCAUCACUCCCCUAUACUCAAUCU .(((((((.(((.((....)).)))...)))))))(((....))).........((((((((..........))))))))................... ( -20.70) >consensus UACAAACUUCGAAAUGGCAAUCUCGUUUAGUUUGUAUCUUCUGGCCCUCGUAACGAUGGCAAACAUGCCACCUUGCCAUCACUCCCCUAUACUCAAUCU ((((((((.(((.((....)).)))...))))))))..................((((((((..........))))))))................... (-18.39 = -18.33 + -0.06)

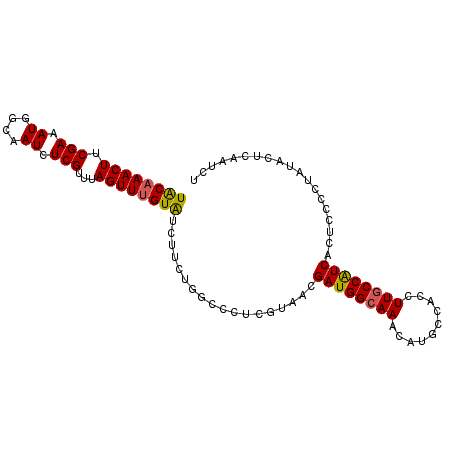
| Location | 17,322,106 – 17,322,205 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
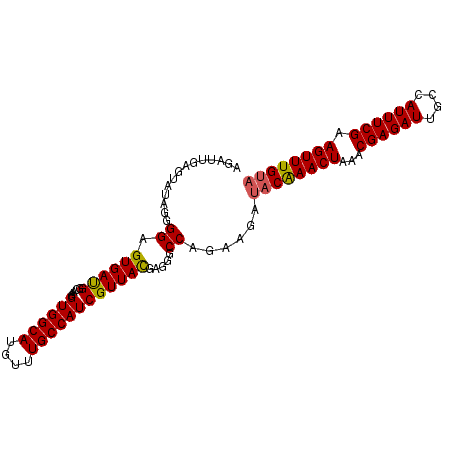
>2L_DroMel_CAF1 17322106 99 - 22407834 AGAUUGAGUAUAGGGGAGUGAUGGCAAGGUGGCAUGUUUGCCAUCGUUAUGAGGGCCAGAAGAUACGAACUAAACGAGAUUGCCAUUUCGAAGUUUGUA .....................((((..(((((((....))))))).........)))).....((((((((...((((((....)))))).)))))))) ( -25.40) >DroSec_CAF1 19763 97 - 1 AGAUUGAGUAUAGGGGAGUGACGGCAAGGUGGCAUGUUUGCCAUCGUUACGA--GCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUA ..............((.((((((.....((((((....))))))))))))..--.))......((((((((...((((((....)))))).)))))))) ( -27.30) >DroSim_CAF1 22111 99 - 1 AGAUUGAGUAUAGGGGAGUGAUGGUAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUA ..............((.....(.(((((((((((....))))))).)))).)...))......((((((((...((((((....)))))).)))))))) ( -26.60) >DroYak_CAF1 14738 99 - 1 AGAUUGAGUAUAGGGGAGUGAUGGCAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGACACAAACUAAACGAGAUUGCUAUUUCGAAGUUUGCA .................((..((((..(((((((....))))))).........))))....)).((((((...((((((....)))))).)))))).. ( -24.20) >consensus AGAUUGAGUAUAGGGGAGUGAUGGCAAGGUGGCAUGUUUGCCAUCGUUACGAGGGCCAGAAGAUACAAACUAAACGAGAUUGCCAUUUCGAAGUUUGUA ..............((.((((((.....((((((....)))))))))))).....))......((((((((...((((((....)))))).)))))))) (-23.49 = -23.42 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:54 2006