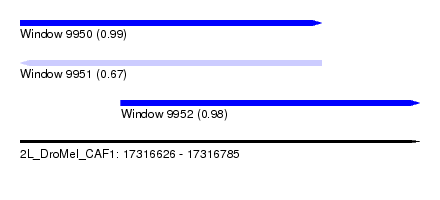
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,316,626 – 17,316,785 |
| Length | 159 |
| Max. P | 0.994186 |

| Location | 17,316,626 – 17,316,746 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -27.58 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17316626 120 + 22407834 UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUACAAAAUUUCGAAACAAAAGUUUGUUUGCCUCAAAGGAGGCGAAAUGUGGCGAAAGGAGGUUUGUAUAAGGUGGGAUUUCGAA ...(((((((.(((((((((.....(((((.((........)).))))).....))))....(((((((....)))))))))))).)))))))(((((((..........)))))))... ( -28.20) >DroSec_CAF1 14216 120 + 1 UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUACAAAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAA ...(((((((.(((((.....................(((((.(((((...))))).)))))(((((((....)))))))))))).)))))))((((((.(........).))))))... ( -34.10) >DroSim_CAF1 15751 120 + 1 UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUACAAAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAA ...(((((((.(((((.....................(((((.(((((...))))).)))))(((((((....)))))))))))).)))))))((((((.(........).))))))... ( -34.10) >DroEre_CAF1 16204 119 + 1 UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUACAAAAUUUUGAAACAAAAGUUUGU-UGCCUCCAAGGAGGCGGAGUGUGGCGAAAGCAGGUUGCUAUAAGGUGGCAAUUCGAA ...(((((((.(((((.(...................(((((.(((((...))))).)))))-.(((((....)))))).))))).)))))))(..((((((((...))))))))..).. ( -36.20) >DroYak_CAF1 8492 120 + 1 UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUAGAAAAUUUUGAAACAAAAGUUUGUUUGCCUCGAAGGAGGCGGAAUGUGGCGAAAGGAGCUUGCUAAAAGGUGGGAUUUCGAA ...(((((((.(((((.((((((((......))).)))).........((((((....))))))(((((....)))))).))))).)))))))((.((..((....))..))...))... ( -34.60) >consensus UUACUUUUGUGGCAUUGCUUACUAAUUGAAUUUAUGUACAAAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAA ...(((((((.(((((...........((((((.(((.............))).))))))..(((((((....)))))))))))).)))))))((((((.(........).))))))... (-27.58 = -28.50 + 0.92)

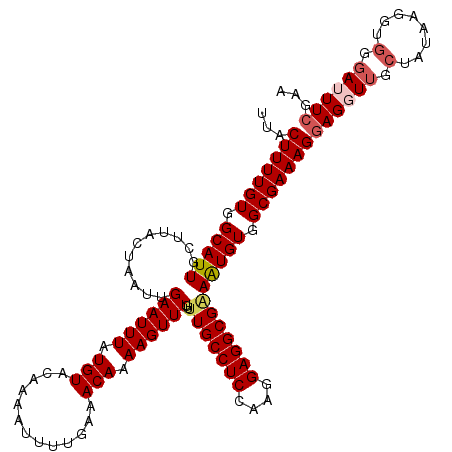
| Location | 17,316,626 – 17,316,746 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -17.45 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.64 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17316626 120 - 22407834 UUCGAAAUCCCACCUUAUACAAACCUCCUUUCGCCACAUUUCGCCUCCUUUGAGGCAAACAAACUUUUGUUUCGAAAUUUUGUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA .................(((........(((((.........(((((....)))))((((((....)))))))))))((((((.(((..................)))..))))))))). ( -15.77) >DroSec_CAF1 14216 120 - 1 UUCGAAAUCCCACCUUAUAGCAACCUCCUUUCGCCACAUUUCGCCUCCUUGGAGGCAAACAAACUUUUGUUUCAAAAUUUUGUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA ..(((((.....................))))).........(((((....)))))......((((((((..((.....((((...(((......)))....))))))..)))))))).. ( -17.20) >DroSim_CAF1 15751 120 - 1 UUCGAAAUCCCACCUUAUAGCAACCUCCUUUCGCCACAUUUCGCCUCCUUGGAGGCAAACAAACUUUUGUUUCAAAAUUUUGUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA ..(((((.....................))))).........(((((....)))))......((((((((..((.....((((...(((......)))....))))))..)))))))).. ( -17.20) >DroEre_CAF1 16204 119 - 1 UUCGAAUUGCCACCUUAUAGCAACCUGCUUUCGCCACACUCCGCCUCCUUGGAGGCA-ACAAACUUUUGUUUCAAAAUUUUGUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA ...................(((...(((((.(..........(((((....))))).-(((((.(((((...))))).)))))...............).))))).)))........... ( -21.60) >DroYak_CAF1 8492 120 - 1 UUCGAAAUCCCACCUUUUAGCAAGCUCCUUUCGCCACAUUCCGCCUCCUUCGAGGCAAACAAACUUUUGUUUCAAAAUUUUCUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA .............(((((.(((.(((................(((((....)))))((((((....)))))).............................)))..)))...)))))... ( -15.50) >consensus UUCGAAAUCCCACCUUAUAGCAACCUCCUUUCGCCACAUUUCGCCUCCUUGGAGGCAAACAAACUUUUGUUUCAAAAUUUUGUACAUAAAUUCAAUUAGUAAGCAAUGCCACAAAAGUAA ..........................................(((((....)))))......((((((((..((........(((.(((......)))))).....))..)))))))).. (-15.80 = -15.64 + -0.16)

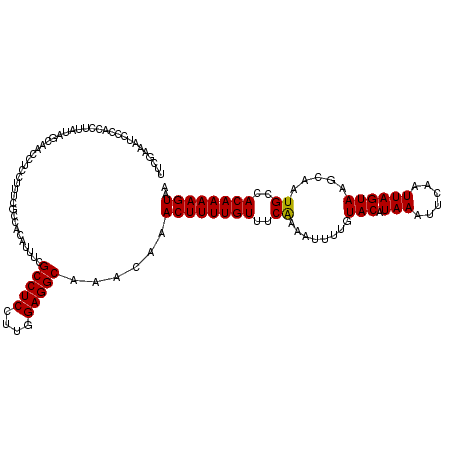
| Location | 17,316,666 – 17,316,785 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
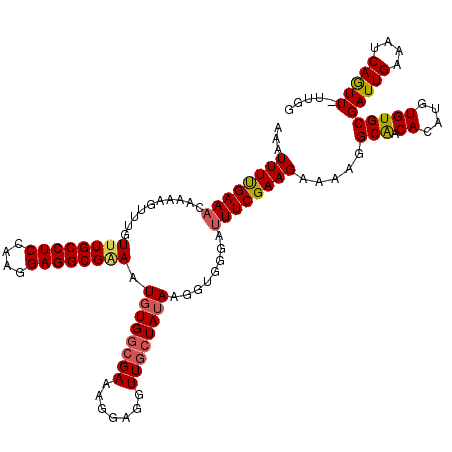
>2L_DroMel_CAF1 17316666 119 + 22407834 AAAUUUCGAAACAAAAGUUUGUUUGCCUCAAAGGAGGCGAAAUGUGGCGAAAGGAGGUUUGUAUAAGGUGGGAUUUCGAAGAAAAGGCAACACAUGUGUGCGAUUGAAAUCAGUU-UUGG ......((((((....(((..((((((((....))))))))....))).......................((((((((.(...(.(((.....))).).)..)))))))).)))-))). ( -27.10) >DroSec_CAF1 14256 119 + 1 AAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAAGAAAAGGCAACACAUGUGUGCGAUUGAAAUCAGUU-UUGG .....(..((((.........((((((((....)))))))).((((((((.......))))))))......((((((((.(...(.(((.....))).).)..)))))))).)))-)..) ( -31.50) >DroSim_CAF1 15791 119 + 1 AAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAAGAAAAGGCAACACAUGUGUGCGAUUGAAAUCAGUU-UUGG .....(..((((.........((((((((....)))))))).((((((((.......))))))))......((((((((.(...(.(((.....))).).)..)))))))).)))-)..) ( -31.50) >DroEre_CAF1 16244 118 + 1 AAAUUUUGAAACAAAAGUUUGU-UGCCUCCAAGGAGGCGGAGUGUGGCGAAAGCAGGUUGCUAUAAGGUGGCAAUUCGAAGAACAGGCGCCACAUGUGUGCGAUUGAAAUCAGUU-CUGG (((((((......))))))).(-((((((....))))))).((((((((....(..((((((((...))))))))..).........))))))))......(((((....)))))-.... ( -35.32) >DroYak_CAF1 8532 120 + 1 AAAUUUUGAAACAAAAGUUUGUUUGCCUCGAAGGAGGCGGAAUGUGGCGAAAGGAGCUUGCUAAAAGGUGGGAUUUCGAAGAACAGGCACCACAUGUGUGCGAUUGAAAUCAAUUUUUGG ...........((((((((.(((((((((....)))))....(((..(((((....((..((....))..)).)))))....))).(((((....).)))).....)))).)))))))). ( -34.30) >consensus AAAUUUUGAAACAAAAGUUUGUUUGCCUCCAAGGAGGCGAAAUGUGGCGAAAGGAGGUUGCUAUAAGGUGGGAUUUCGAAGAAAAGGCAACACAUGUGUGCGAUUGAAAUCAGUU_UUGG ...((((((((..........((((((((....)))))))).((((((((.......))))))))........)))))))).....(((.((....)))))(((((....)))))..... (-26.40 = -26.80 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:47 2006