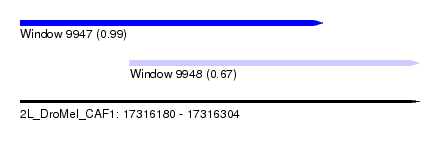
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,316,180 – 17,316,304 |
| Length | 124 |
| Max. P | 0.994975 |

| Location | 17,316,180 – 17,316,274 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
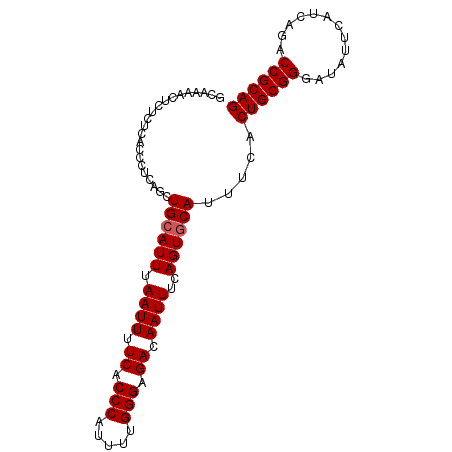
>2L_DroMel_CAF1 17316180 94 + 22407834 GCAAAACUCUCUCACCCUCAGCUGCAUUUAAUUUUCACCCAUUUUGGGAGACAAUUUCAGUGCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ......................((((((.((((.((.(((.....))).)).))))..)))))).....((((((.............)))))) ( -22.02) >DroSec_CAF1 13770 94 + 1 GCAAAACUCUCUCACCCUCAGCUGCAUUUAAUUUUCACCCAUUUUGGGAGACAAUUUCAGUCCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ....................((((.....((((.((.(((.....))).)).)))).))))........((((((.............)))))) ( -18.02) >DroSim_CAF1 15299 94 + 1 GCAAAACUCUCUCACCCUCAGCUGCAUUUAAUUUUCACCCAUUUUGGGAGACAAUUUCAGUGCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ......................((((((.((((.((.(((.....))).)).))))..)))))).....((((((.............)))))) ( -22.02) >DroEre_CAF1 15769 91 + 1 GCAAACCUCAC---CCCCCGGCUGCAUUUAAUUUUCGCCCAUUUUGGGAGACAAUUUCAGUGCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ...........---........((((((.((((.((.(((.....))).)).))))..)))))).....((((((.............)))))) ( -22.52) >DroYak_CAF1 8081 91 + 1 GCAAAGCUCUC---CCCUCCGCUGCAUUUAAUUUUCACCCAUUUUGGGAGACAAUUUCAGUGCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ....(((....---......)))(((((.((((.((.(((.....))).)).))))..)))))......((((((.............)))))) ( -22.32) >consensus GCAAAACUCUCUCACCCUCAGCUGCAUUUAAUUUUCACCCAUUUUGGGAGACAAUUUCAGUGCAUUUCACUGCGGGAUAUUCAUCAGACCGCAG ......................((((((.((((.((.(((.....))).)).))))..)))))).....((((((.............)))))) (-20.82 = -21.02 + 0.20)

| Location | 17,316,214 – 17,316,304 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
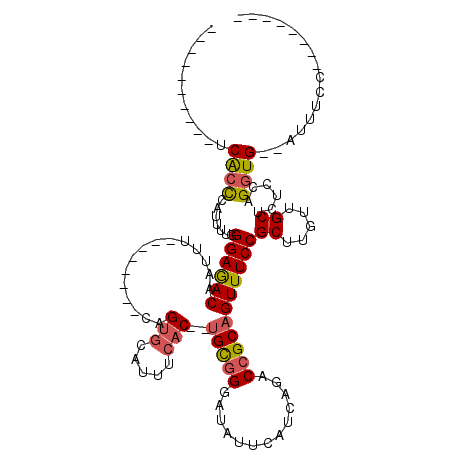
>2L_DroMel_CAF1 17316214 90 + 22407834 ----------UCACCCAUUUUGGGAGACAAUUU--------CAGUGCAUUUCAC--UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGUUGCUCUCCAGGUG--AUUUCC-------- ----------(((((.....((((((.((((..--------.((((......((--(((((.............)))))))...)))).))))))).)))))))--).....-------- ( -28.12) >DroSec_CAF1 13804 90 + 1 ----------UCACCCAUUUUGGGAGACAAUUU--------CAGUCCAUUUCAC--UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGUUGCUCUCCAGGUG--AUUUCG-------- ----------(((((.....((((((.((((..--------.(((.......((--(((((.............)))))))....))).))))))).)))))))--).....-------- ( -25.82) >DroSim_CAF1 15333 90 + 1 ----------UCACCCAUUUUGGGAGACAAUUU--------CAGUGCAUUUCAC--UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGUUGCUCUCCAGGUG--AUUUCC-------- ----------(((((.....((((((.((((..--------.((((......((--(((((.............)))))))...)))).))))))).)))))))--).....-------- ( -28.12) >DroEre_CAF1 15800 98 + 1 ----------UCGCCCAUUUUGGGAGACAAUUU--------CAGUGCAUUUCAC--UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGCUGCUCUCCAGGUG--AUUUCCCCUCUUUC ----------((.(((.....))).))......--------(((((.....)))--)).((((...(((((.((..(((((........)))))..))..))))--)..))))....... ( -28.20) >DroYak_CAF1 8112 90 + 1 ----------UCACCCAUUUUGGGAGACAAUUU--------CAGUGCAUUUCAC--UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGUUGCUCUCCAGGUG--AUUUCC-------- ----------(((((.....((((((.((((..--------.((((......((--(((((.............)))))))...)))).))))))).)))))))--).....-------- ( -28.12) >DroAna_CAF1 23771 112 + 1 AUUGAUUGGAUCACUCGGUUAGGGAAACGGUUUCUGGCCCUCAGUGCACUUCCCCGUGUGGCAUAUUCAUCAGGCAGCCGUUUCCGCUUGUUGCUCUUCGGUUGCGGCCUCG-------- .((((((((.....))))))))(((((((((((((((......((((((........)).)))).....))))).))))))))))....(((((.(....)..)))))....-------- ( -34.00) >consensus __________UCACCCAUUUUGGGAGACAAUUU________CAGUGCAUUUCAC__UGCGGGAUAUUCAUCAGACCGCAGUUUCCGCUUGUUGCUCUCCAGGUG__AUUUCC________ ...........((((.......((((((...............(((.....)))..(((((.............)))))))))))((.....))......))))................ (-16.76 = -17.04 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:43 2006