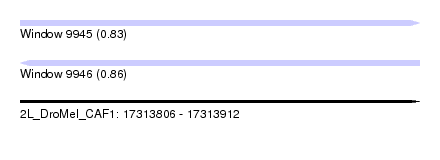
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,313,806 – 17,313,912 |
| Length | 106 |
| Max. P | 0.858776 |

| Location | 17,313,806 – 17,313,912 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
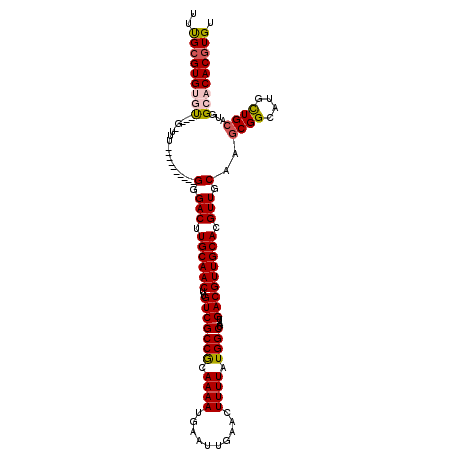
>2L_DroMel_CAF1 17313806 106 + 22407834 UUUGCGUGUGU---G-UUU----------GGGACUUGCAACUUGUCGCCGCAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAAGCGGCAUGCUGCAUGGCACACGUGU ...((((((((---.-.((----------(.(((.((((((..((((..((...(((((.......))))).))...)))))))))).))).)))((((....))))...)))))))).. ( -37.50) >DroSec_CAF1 11438 106 + 1 UUUGCGUGUGU---G-UUU----------GGGACUUGCAACUUGUCGCCGCAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAAGCGGCAUGCUGCAUGGCACACGUGU ...((((((((---.-.((----------(.(((.((((((..((((..((...(((((.......))))).))...)))))))))).))).)))((((....))))...)))))))).. ( -37.50) >DroSim_CAF1 12914 106 + 1 UUUGCGUGUGU---G-UUU----------GGGACUUGCAACUUGUCGCCGCAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAAGCGGCAUGCUGCAUGGCACACGUGU ...((((((((---.-.((----------(.(((.((((((..((((..((...(((((.......))))).))...)))))))))).))).)))((((....))))...)))))))).. ( -37.50) >DroEre_CAF1 13685 104 + 1 UUUGCGUG--U---G-UUU----------GGGACUUGCAACUUGUCGCCGCAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAAGCGGCAUGUUGCAUGGCACACGUGU ...(((((--(---(-((.----------......((((((.((((((.((...(((((.......))))).))........((((.....)))))))))).)))))).))))))))).. ( -35.80) >DroAna_CAF1 21539 120 + 1 UGCGAGUGGCCGAAGCUUUAAGGGACGAGGGGACCUGCAACUUGUCGCCACAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAUACGGCAUGUUGCGUGGCGCACGUGU .(((.(((((((........(((..(....)..)))(((((.((((((((.((((..........)))).)))).......(((((.....))).)))))).))))).)))).))).))) ( -35.60) >consensus UUUGCGUGUGU___G_UUU__________GGGACUUGCAACUUGUCGCCGCAAAAUGAAUUGAACUUUUAUGGCUAAUGACGUUGCACGUUGCAAGCGGCAUGCUGCAUGGCACACGUGU ..(((((((((..................(.(((.((((((..(((((((.((((..........)))).))))....))))))))).))).)..((((....))))...))))))))). (-28.40 = -28.88 + 0.48)

| Location | 17,313,806 – 17,313,912 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
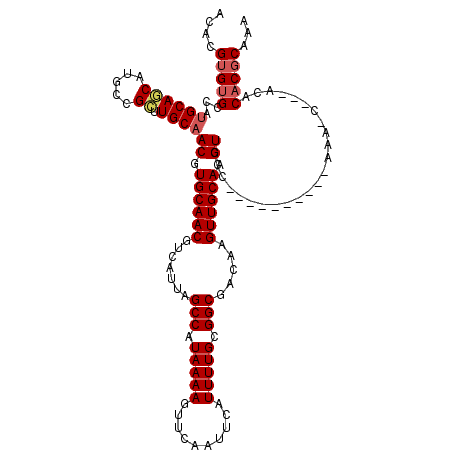
>2L_DroMel_CAF1 17313806 106 - 22407834 ACACGUGUGCCAUGCAGCAUGCCGCUUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGCGGCGACAAGUUGCAAGUCCC----------AAA-C---ACACACGCAAA ...((((((...((((((.....)).))))((.((((((.......(((.(((((..........))))).))).....)))))).))...----------...-.---.)))))).... ( -28.10) >DroSec_CAF1 11438 106 - 1 ACACGUGUGCCAUGCAGCAUGCCGCUUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGCGGCGACAAGUUGCAAGUCCC----------AAA-C---ACACACGCAAA ...((((((...((((((.....)).))))((.((((((.......(((.(((((..........))))).))).....)))))).))...----------...-.---.)))))).... ( -28.10) >DroSim_CAF1 12914 106 - 1 ACACGUGUGCCAUGCAGCAUGCCGCUUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGCGGCGACAAGUUGCAAGUCCC----------AAA-C---ACACACGCAAA ...((((((...((((((.....)).))))((.((((((.......(((.(((((..........))))).))).....)))))).))...----------...-.---.)))))).... ( -28.10) >DroEre_CAF1 13685 104 - 1 ACACGUGUGCCAUGCAACAUGCCGCUUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGCGGCGACAAGUUGCAAGUCCC----------AAA-C---A--CACGCAAA ...((((((....((.....)).((((((((((.....)(((....(((.(((((..........))))).))))))..)))))))))...----------...-)---)--)))).... ( -27.20) >DroAna_CAF1 21539 120 - 1 ACACGUGCGCCACGCAACAUGCCGUAUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGUGGCGACAAGUUGCAGGUCCCCUCGUCCCUUAAAGCUUCGGCCACUCGCA ....(((.(((..(((((.((((((.....))).)))..(((....(((((((((..........))))))))))))..)))))(((..(....)..))).........))))))..... ( -35.50) >consensus ACACGUGUGCCAUGCAGCAUGCCGCUUGCAACGUGCAACGUCAUUAGCCAUAAAAGUUCAAUUCAUUUUGCGGCGACAAGUUGCAAGUCCC__________AAA_C___ACACACGCAAA ....(((((...((((((.....)).))))((.((((((.......(((.(((((..........))))).))).....)))))).))........................)))))... (-22.26 = -22.82 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:41 2006