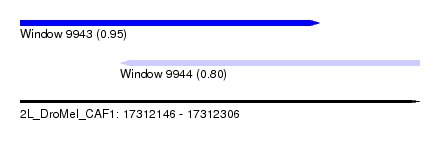
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,312,146 – 17,312,306 |
| Length | 160 |
| Max. P | 0.952224 |

| Location | 17,312,146 – 17,312,266 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.95 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
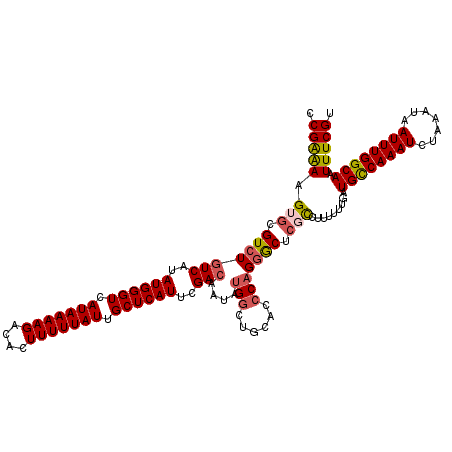
>2L_DroMel_CAF1 17312146 120 + 22407834 CCGAAAAGUGCGUCUGUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGCUGCACCCAGGACUCUCCUUUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((.(((((((((((..((((((.(((((((....))))))).))))))..))))....))).))))..(((.....))).......((((((((.......)))))))).))))). ( -34.20) >DroSec_CAF1 9786 120 + 1 CCGAAAAGUGCGUCUGUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGCUGCACCCAGGGCUCGCCUUUUUUGAUGUCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((.(((((((((((..((((((.(((((((....))))))).))))))..))))....))).))))..((((....))))......((((((((.......)))))))).))))). ( -32.40) >DroSim_CAF1 11272 120 + 1 CCGAAAAGUGCGUCUUUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGGUGCACCCAGGGCUCGCCUUUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((.(((.((((.((..((((((.(((((((....))))))).))))))..)).....((((....)))))))).))).........((((((((.......)))))))).))))). ( -33.40) >DroEre_CAF1 12138 120 + 1 CCGAAAAGUGCGUCUGUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGCUGCACCCUGGGCCCGCCCUUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((((.(((..((((..((((((.(((((((....))))))).))))))..))))....((((........))))))).))))))).((((((((.......))))))))....... ( -37.30) >DroYak_CAF1 4603 120 + 1 CCGAAAAGUGCGUCUGUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGCUGCACCCAGGGCUCGCCUUUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((.(((((((((((..((((((.(((((((....))))))).))))))..))))....))).))))..((((....))))......((((((((.......)))))))).))))). ( -35.10) >DroAna_CAF1 20356 114 + 1 CCGGGAAG--CGUCUGUCAUAUGGGUCAUAAAAGCCAGUUUUUAUUGCUCAUCUGACAAUAUGG----ACGCAGAGCCCGUCGCUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .((((..(--(((((((((.((((((.(((((((....))))))).)))))).)))).....))----))))....))))..........((((((((.......))))))))....... ( -38.90) >consensus CCGAAAAGUGCGUCUGUCAUAUGGGUCAUAAAAGACACUUUUUAUUGCUCAUUCGACAAUAUGGCUGCACCCAGGGCUCGCCUUUUUUGAUGCCAAAUCUAAAUAAUUUGGCAAUUUCGU .(((((.(((.(((((((..((((((.(((((((....))))))).))))))..)))....(((......))))))).))).........((((((((.......)))))))).))))). (-28.48 = -28.95 + 0.47)

| Location | 17,312,186 – 17,312,306 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.97 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
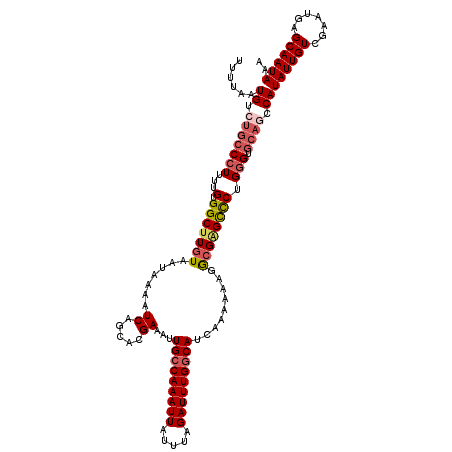
>2L_DroMel_CAF1 17312186 120 - 22407834 AUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAAAGGAGAGUCCUGGGUGCAGCCAUAUUGUCGAAUGAGCAAUAAA ......((.(((((((((((((.((.((......)).))))))))..(((((((((.....))))))))).......(((.....)))))).)))).))((((((.......)))))).. ( -29.80) >DroSec_CAF1 9826 120 - 1 UUUUAAUGUCUGCCCUUUUGUGGCCUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGACAUCAAAAAAGGCGAGCCCUGGGUGCAGCCAUAUUGUCGAAUGAGCAAUAAA ......((.(((((((...((.((((............(((......)))((((((.....))))))..........))))..))...))).)))).))((((((.......)))))).. ( -24.90) >DroSim_CAF1 11312 120 - 1 UUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAAAGGCGAGCCCUGGGUGCACCCAUAUUGUCGAAUGAGCAAUAAA ......((..((((((...(.(((((((.......((.....))...(((((((((.....))))))))).........)))))))).))).)))..))((((((.......)))))).. ( -31.80) >DroEre_CAF1 12178 120 - 1 UUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAAGGGCGGGCCCAGGGUGCAGCCAUAUUGUCGAAUGAGCAAUAAA .......(((((((((((((((.((.((......)).))))).....(((((((((.....)))))))))...))))))))))))....((.....)).((((((.......)))))).. ( -36.20) >DroYak_CAF1 4643 120 - 1 UUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAAAGGCGAGCCCUGGGUGCAGCCAUAUUGUCGAAUGAGCAAUAAA ......((.(((((((...(.(((((((.......((.....))...(((((((((.....))))))))).........)))))))).))).)))).))((((((.......)))))).. ( -34.10) >DroAna_CAF1 20394 116 - 1 UUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAGCGACGGGCUCUGCGU----CCAUAUUGUCAGAUGAGCAAUAAA .....((((..(((((((((((.((.((......)).))))))))).(((((((((.....)))))))))...........))))...))))----...(((((((....)).))))).. ( -28.60) >consensus UUUUAAUGUCUGCCCUUUUGUGGCUUGUAAUAAAAUCAGCACGAAAUUGCCAAAUUAUUUAGAUUUGGCAUCAAAAAAGGCGAGCCCUGGGUGCAGCCAUAUUGUCGAAUGAGCAAUAAA ......((.(((((((...(.(((((((.......((.....))...(((((((((.....))))))))).........)))))))).))).)))).))((((((.......)))))).. (-26.05 = -26.97 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:39 2006