| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,309,808 – 17,309,919 |
| Length | 111 |
| Max. P | 0.678553 |

| Location | 17,309,808 – 17,309,919 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -37.21 |
| Consensus MFE | -24.13 |
| Energy contribution | -26.68 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
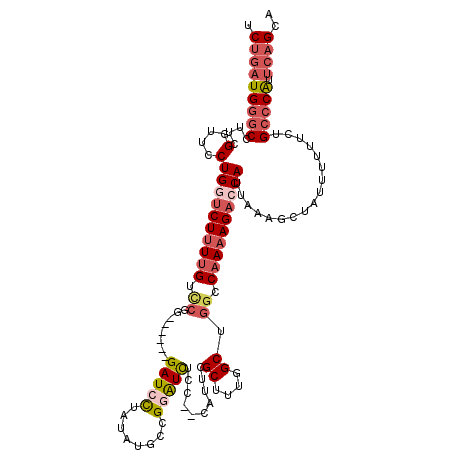
>2L_DroMel_CAF1 17309808 111 + 22407834 UGCUGAAUGGGCAGAAAAAAUAGCUUUAAUGGUCUUUUGGCCAGCCAAAGCGAAUA--GGAGAUCCGGCAUAUAGGAUC------CCGGACAAAAGACCAGGAACCGAGGG-CCAUCAGA ..((((.(((.(.................((((((((((.((.((....)).....--((.(((((........)))))------)))).))))))))))(....)...).-))))))). ( -38.00) >DroSec_CAF1 7434 112 + 1 UGCUGAAUGGGCAGAAAAAAUAGCUUUAAUGGUCUUUUGGCCAGCCAAAGCGAAUA--GGAGAUCCGGCAUAUAGGAUC------CCGGACAAAAGACCAGGAACCGAAGGCCCAUCAGA ..((((.(((((.................((((((((((.((.((....)).....--((.(((((........)))))------)))).))))))))))(....)....))))))))). ( -42.60) >DroSim_CAF1 8920 112 + 1 UGCUGAAUGGGCAGAAAAAAUAUCUUUAAUGGUCUUUUGGCCAGCCAAAGCGAAUA--GGAGAUCCGGCAUAUAGGAUC------CCGGACAAAAGAGCAGGAACCGAAGGCCCAUCAGA ..((((.(((((.................((.(((((((.((.((....)).....--((.(((((........)))))------)))).))))))).))(....)....))))))))). ( -38.10) >DroEre_CAF1 9928 111 + 1 UGCUGAAUGGGCAGAAAAAAUAGCUUUAAUGGUCUUUUGGCCAGCCAAAGCGAAUG--GGGAACACGGAACAUAGGAUC------CCGGACAAAAGACCAGGAACCGA-GGCCCAUCAGA ..((((.(((((.................((((((((((.((.((....)).....--((((.(.(........)).))------)))).))))))))))(....)..-.))))))))). ( -37.80) >DroYak_CAF1 2282 112 + 1 UGCUGAAUGGGCAGAAAAAAAAGCUUUAAUGGUCUUUUGGGUAGCCAAAGCGAAUG--GGAGAUCCGGAAUACAGGAUC------CCGGACAAAAGACCAGGAACCGAAGGCCCAUCAGA ..((((.(((((.................((((((((((....((....)).....--((.(((((........)))))------))...))))))))))(....)....))))))))). ( -39.30) >DroAna_CAF1 18391 118 + 1 UGCUGAACGGGCAGAAAAAAUAUUUUUAAUGGUCUUUUGGCCAGC-AAAGCGGACGAAGAGGAUACAAAGGACGAGAUCCAGUGCCAGAACUACAGG-CAGGAAGCCAAGGCGCAAGCGA .........(((...................((((((((.((...-..............))...))))))))....(((..((((.........))-))))).)))...((....)).. ( -27.43) >consensus UGCUGAAUGGGCAGAAAAAAUAGCUUUAAUGGUCUUUUGGCCAGCCAAAGCGAAUA__GGAGAUCCGGAAUAUAGGAUC______CCGGACAAAAGACCAGGAACCGAAGGCCCAUCAGA ..((((.(((((.................((((((((((.((.((....))..........(((((........)))))........)).))))))))))(....)....))))))))). (-24.13 = -26.68 + 2.56)


| Location | 17,309,808 – 17,309,919 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -22.88 |
| Energy contribution | -24.55 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17309808 111 - 22407834 UCUGAUGG-CCCUCGGUUCCUGGUCUUUUGUCCGG------GAUCCUAUAUGCCGGAUCUCC--UAUUCGCUUUGGCUGGCCAAAAGACCAUUAAAGCUAUUUUUUCUGCCCAUUCAGCA .((((.((-(....(....)((((((((((.((((------(((((........))))))).--.....((....)).)).)))))))))).................)))...)))).. ( -34.00) >DroSec_CAF1 7434 112 - 1 UCUGAUGGGCCUUCGGUUCCUGGUCUUUUGUCCGG------GAUCCUAUAUGCCGGAUCUCC--UAUUCGCUUUGGCUGGCCAAAAGACCAUUAAAGCUAUUUUUUCUGCCCAUUCAGCA .((((((((((....)....((((((((((.((((------(((((........))))))).--.....((....)).)).)))))))))).................))))).)))).. ( -40.70) >DroSim_CAF1 8920 112 - 1 UCUGAUGGGCCUUCGGUUCCUGCUCUUUUGUCCGG------GAUCCUAUAUGCCGGAUCUCC--UAUUCGCUUUGGCUGGCCAAAAGACCAUUAAAGAUAUUUUUUCUGCCCAUUCAGCA .(((((((((..........((.(((((((.((((------(((((........))))))).--.....((....)).)).))))))).))....(((.......)))))))).)))).. ( -34.20) >DroEre_CAF1 9928 111 - 1 UCUGAUGGGCC-UCGGUUCCUGGUCUUUUGUCCGG------GAUCCUAUGUUCCGUGUUCCC--CAUUCGCUUUGGCUGGCCAAAAGACCAUUAAAGCUAUUUUUUCUGCCCAUUCAGCA .(((((((((.-..((.((((((.(....).))))------)).))................--.....(((((((.(((........))))))))))..........))))).)))).. ( -34.90) >DroYak_CAF1 2282 112 - 1 UCUGAUGGGCCUUCGGUUCCUGGUCUUUUGUCCGG------GAUCCUGUAUUCCGGAUCUCC--CAUUCGCUUUGGCUACCCAAAAGACCAUUAAAGCUUUUUUUUCUGCCCAUUCAGCA .((((((((((....)....(((((((((((((((------(((.....))))))))....(--((.......))).....)))))))))).................))))).)))).. ( -37.50) >DroAna_CAF1 18391 118 - 1 UCGCUUGCGCCUUGGCUUCCUG-CCUGUAGUUCUGGCACUGGAUCUCGUCCUUUGUAUCCUCUUCGUCCGCUUU-GCUGGCCAAAAGACCAUUAAAAAUAUUUUUUCUGCCCGUUCAGCA ..(((.((.....(((.....)-))....))...)))...((((..((.....)).)))).........(((..-((.(((..(((((.............)))))..))).))..))). ( -22.02) >consensus UCUGAUGGGCCUUCGGUUCCUGGUCUUUUGUCCGG______GAUCCUAUAUGCCGGAUCUCC__CAUUCGCUUUGGCUGGCCAAAAGACCAUUAAAGCUAUUUUUUCUGCCCAUUCAGCA .(((((((((....(....)((((((((((.((........(((((........)))))..........((....)).)).)))))))))).................))))).)))).. (-22.88 = -24.55 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:32 2006