| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,308,872 – 17,308,991 |
| Length | 119 |
| Max. P | 0.650534 |

| Location | 17,308,872 – 17,308,991 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -16.98 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
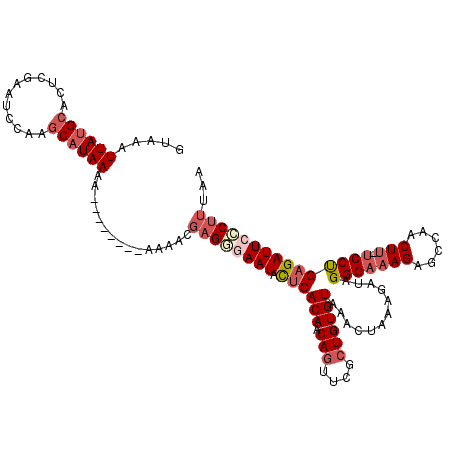
>2L_DroMel_CAF1 17308872 119 + 22407834 GUAAAUUAUGCACUCGAAUCCAAGCAUAAAAAAAAAAAAAAAACGAGGGAAUACUCACAACAGUUCGCUGUGUAAACUAAAGAUAGGGAAAGAGCCAACUU-UCCUGAGAUUCCCUUUAA .....((((((............))))))...............((((((((.((((((.(((....))))))............(((((((......)))-)))))))))))))))... ( -30.60) >DroSec_CAF1 6390 111 + 1 GUAAAUUAUGCACCCGAAUCCAAGCAUAAAA--------AAAACGAGGGAAUACUCACAACAGUUCGCUGUGUAAACUAAAGAUAGGGAAAGAGCCAACUU-UCCUGAGAUUCCCUUUAA .....((((((............))))))..--------.....((((((((.((((((.(((....))))))............(((((((......)))-)))))))))))))))... ( -30.60) >DroSim_CAF1 7883 111 + 1 GUAAAUUAUGCACUCGAAUCCAAGCAUAAAA--------AAAACGAGGGAAUACUCACAACAGUUCGCUGUGUAAACUAAAGAUAGGGAAAGAGCCAACUU-UCCUGAGAUUCCCUUUAA .....((((((............))))))..--------.....((((((((.((((((.(((....))))))............(((((((......)))-)))))))))))))))... ( -30.60) >DroEre_CAF1 9050 111 + 1 GUAAAUUAUGCACUCGAAUCCAAGCAUAAAA---------AAACGAGCGAAUACUCACAACAGUUCGCUGUGUAAACUUAAGAUAGGGAAAGAGCCAACUUUUCCCGAGAUUCCCUUUAA .....((((((............))))))..---------....(((.((((.((((((.(((....))))))............(((((((.......)))))))))))))).)))... ( -23.80) >DroYak_CAF1 1341 111 + 1 GUAAAUUAUGCAGUCGAAUCCAAGCAUAAAA---------AAACGAGAAAAUACUCACAACAUUUCGCUGUGUAAACUAAAGAUAGGGAAAGAGCCAACUUUUCCUGAGAUUCCCUUUAA .....((((((((.((((.....(.......---------...)(((......))).......))))))))))))........(((((((((......)))))))))............. ( -19.70) >DroAna_CAF1 17530 94 + 1 GUAAAUUAUGCACCCGAAAC----CCUAAAA--------AAAA-CUGAAAAUAUCCACAACAGUCCUCUGUGUAAACAAAAAACGGGGGG-------------CCUGAGAUUCUCUUUAA ....................----.......--------....-................(((((((((((...........))))))))-------------.)))............. ( -13.40) >consensus GUAAAUUAUGCACUCGAAUCCAAGCAUAAAA________AAAACGAGGGAAUACUCACAACAGUUCGCUGUGUAAACUAAAGAUAGGGAAAGAGCCAACUU_UCCUGAGAUUCCCUUUAA .....((((((............))))))...............((((((((.((((((.(((....))))))............(((((((......))).)))))))))))))))... (-16.98 = -18.62 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:30 2006