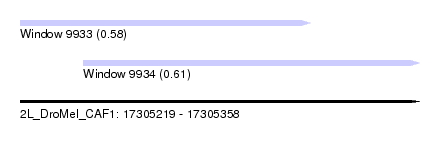
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,305,219 – 17,305,358 |
| Length | 139 |
| Max. P | 0.605378 |

| Location | 17,305,219 – 17,305,320 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
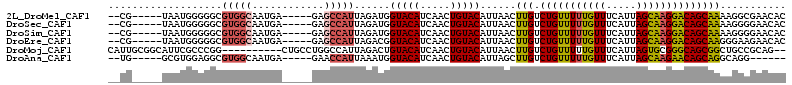
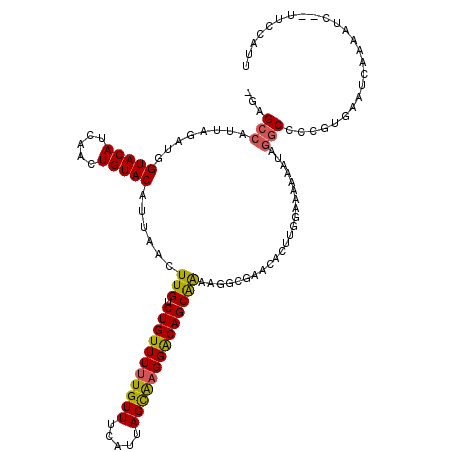
>2L_DroMel_CAF1 17305219 101 + 22407834 --CG-----UAAUGGGGGCGUGGCAAUGA-----GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGCGAACAC --((-----(((((...(((((((.....-----..)))))..((((...))))....)).))))...(((.(((((((((((.....))))))))))))))...)))..... ( -25.50) >DroSec_CAF1 2749 101 + 1 --CG-----UAAUGGGGGCGUGGCAAUGA-----GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGGGAACAC --..-----(((((...(((((((.....-----..)))))..((((...))))....)).)))))..(((.(((((((((((.....))))))))))))))........... ( -24.90) >DroSim_CAF1 3058 101 + 1 --CG-----UAAUGGGGGCGUGGCAAUGA-----GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGGGAACAC --..-----(((((...(((((((.....-----..)))))..((((...))))....)).)))))..(((.(((((((((((.....))))))))))))))........... ( -24.90) >DroEre_CAF1 5580 101 + 1 --CG-----UAAUGGGGGCGUGGCAAUGA-----GAGCCAUUAGACGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAGGGAAGAACAC --..-----(((((.....(((((.....-----..)))))...(((((......))))).))))).((((.(((((((((((.....))))))))))))))).......... ( -29.00) >DroMoj_CAF1 20672 101 + 1 CAUUGCGGCAUUCGCCCGG----------CUGCCUGGCCAUUAGACUGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGUGCGGGCAGCGGCUGCCGCAG-- ...(((((((...(((..(----------(((((((..((((((.....(((..(((.(.(((......)))).)))..)))....)))))))))))))))))))))))).-- ( -38.70) >DroAna_CAF1 14577 95 + 1 --UG-----GCGUGGAGGCGUGGCAAUGA-----GAACCAUUAAAUGGUACAUCAACUGUACAUUAGCUUGUCUGUUUUUGUUUCAUUAGCAAGAACAGCAGGCAGG------ --..-----..(((.((((...))..(((-----..(((((...)))))...))).)).)))....(((((.(((((((((((.....))))))))))))))))...------ ( -29.50) >consensus __CG_____UAAUGGGGGCGUGGCAAUGA_____GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGGGAACAC ...................(((((............)))))......(((((.....)))))......(((.(((((((((((.....))))))))))))))........... (-17.53 = -17.78 + 0.25)
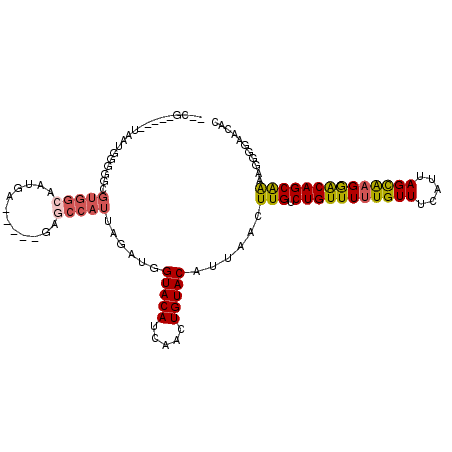
| Location | 17,305,241 – 17,305,358 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17305241 117 + 22407834 -GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGCGAACACUUGGAAAAAAUAGGCCCUGUGAAUCAAAUUC--UUCCAUU -(((((........(((((.....)))))......(((.(((((((((((.....))))))))))))))..)))...(((..((..........))..)))..))......--....... ( -26.80) >DroSec_CAF1 2771 119 + 1 -GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGGGAACACUUGGAAAAAAUAUGCCGCAUGAAUCAAAAUUUAUUGCAUU -...(((..((.(((((((.....))))).((..(((..(((((((((((.....)))))))))))...)))..)).)))))))............((((((((....))))).)))... ( -24.60) >DroSim_CAF1 3080 117 + 1 -GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGGGAACACUUGGAAAAAAUAGGCCGCUUGAAUCAUAAUC--UUGCAUU -(.(((........(((((.....))))).((..(((..(((((((((((.....)))))))))))...)))..))................))))((..((.......))--..))... ( -26.00) >DroEre_CAF1 5602 115 + 1 -GAGCCAUUAGACGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAGGGAAGAACACUUGAAAAAA--AGGCCUCUUGAAUCCUAACC--UUCAAGU -..(((........(((((.....))))).((..((((.(((((((((((.....)))))))))))))))..))...............--.)))..((((((........--)))))). ( -30.20) >DroMoj_CAF1 20695 97 + 1 CUGGCCAUUAGACUGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGUGCGGGCAGCGGCUGCCGCAG------CAGCUGUAGCCUCUGCAAA----------------- ............((((((.........(((..(((......)))..))).......))))))((((.((((((.((..------..)).)))))).))))...----------------- ( -30.69) >consensus _GAGCCAUUAGAUGGUACAUCAACUGUACAUUAACUUGUCUGUUUUUGUUUCAUUAGCAAGGACAGCAAAAGGCGAACACUUGGAAAAAAUAGGCCCCGUGAAUCAAAAUC__UUCCAUU ...(((........(((((.....)))))......(((.(((((((((((.....)))))))))))))).......................)))......................... (-18.14 = -17.94 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:28 2006