
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,302,798 – 17,302,890 |
| Length | 92 |
| Max. P | 0.975476 |

| Location | 17,302,798 – 17,302,890 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -9.27 |
| Energy contribution | -8.65 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17302798 92 + 22407834 GGCAAAUGCAAAUUGCAAUAGCAGCCC---CUACUAAUGCAAUUCCGCG--------------CACUUCGCCCCUCCC-----AGGCAUCCGAAUACCCCUCAACAUAAAGCCG (((....((.(((((((.(((.((...---)).))).)))))))..)).--------------......(((......-----.))).......................))). ( -19.60) >DroSec_CAF1 360 97 + 1 GGCAAAUGCAAAUUGCAAUAGCAGCCC---CUACUAAUGCAAUUCCGCG--------------CACUUUCCCCCUUCCCCGUCGGGCAUUCGAAUACCCCUCAACAUAAAGCCG (((.(((((.(((((((.(((.((...---)).))).)))))))(((((--------------................)).)))))))).((.......))........))). ( -19.39) >DroEre_CAF1 3203 85 + 1 GGCAAAUGCAAAUUGCAAUAGCAGCCC---CUACUAAUGCAAUUCCGCG--------------CACUCCCCC-----C-----UGCCAUCCGAGUGCCCCU--ACAUAAAGCCG (((....((.(((((((.(((.((...---)).))).)))))))..))(--------------(((((....-----.-----........))))))....--.......))). ( -23.22) >DroAna_CAF1 12128 105 + 1 ACCAAAUGGAAAUUGCAACAGAUUUCGCAGAAACUAAUGCAAUACUGCUACGGCGGAGGCUGCCACCCCACC-----C-UAU-UCACACCCCAAUUGCCGG--CCAUAAUGCCG ..(((.(((..((((((..((.((((...))))))..)))))).((((....))))(((.((......)).)-----)-)..-.......))).))).(((--(......)))) ( -24.40) >consensus GGCAAAUGCAAAUUGCAAUAGCAGCCC___CUACUAAUGCAAUUCCGCG______________CACUCCCCC_____C_____UGGCAUCCGAAUACCCCU__ACAUAAAGCCG .((....)).(((((((.(((.((......)).))).)))))))...................................................................... ( -9.27 = -8.65 + -0.62)


| Location | 17,302,798 – 17,302,890 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
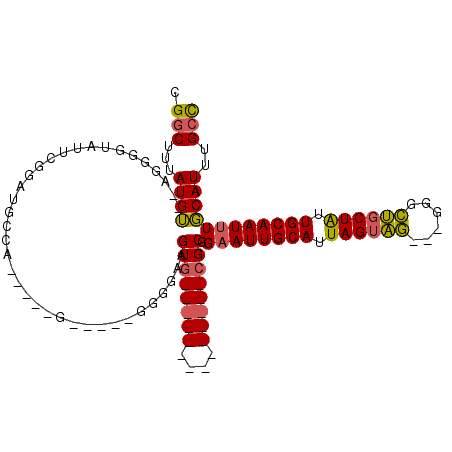
>2L_DroMel_CAF1 17302798 92 - 22407834 CGGCUUUAUGUUGAGGGGUAUUCGGAUGCCU-----GGGAGGGGCGAAGUG--------------CGCGGAAUUGCAUUAGUAG---GGGCUGCUAUUGCAAUUUGCAUUUGCC ((((.....))))..((((((....))))))-----......(((.(((((--------------(...((((((((.((((((---...)))))).))))))))))))))))) ( -31.00) >DroSec_CAF1 360 97 - 1 CGGCUUUAUGUUGAGGGGUAUUCGAAUGCCCGACGGGGAAGGGGGAAAGUG--------------CGCGGAAUUGCAUUAGUAG---GGGCUGCUAUUGCAAUUUGCAUUUGCC ...((((.((((...((((((....))))))))))..))))..((.(((((--------------(...((((((((.((((((---...)))))).)))))))))))))).)) ( -31.80) >DroEre_CAF1 3203 85 - 1 CGGCUUUAUGU--AGGGGCACUCGGAUGGCA-----G-----GGGGGAGUG--------------CGCGGAAUUGCAUUAGUAG---GGGCUGCUAUUGCAAUUUGCAUUUGCC .(((...((((--..(.((((((........-----.-----....)))))--------------).).((((((((.((((((---...)))))).))))))))))))..))) ( -31.82) >DroAna_CAF1 12128 105 - 1 CGGCAUUAUGG--CCGGCAAUUGGGGUGUGA-AUA-G-----GGUGGGGUGGCAGCCUCCGCCGUAGCAGUAUUGCAUUAGUUUCUGCGAAAUCUGUUGCAAUUUCCAUUUGGU ((((......)--))).(((.(((((.....-...-.-----((((((((....))).)))))(((((((..(((((........)))))...)))))))...))))).))).. ( -38.90) >consensus CGGCUUUAUGU__AGGGGUAUUCGGAUGCCA_____G_____GGGGAAGUG______________CGCGGAAUUGCAUUAGUAG___GGGCUGCUAUUGCAAUUUGCAUUUGCC .(((...((((.....................................((((((((....)))))))).((((((((.((((((......)))))).))))))))))))..))) (-18.14 = -18.83 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:22 2006