| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,301,676 – 17,301,767 |
| Length | 91 |
| Max. P | 0.626783 |

| Location | 17,301,676 – 17,301,767 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17301676 91 + 22407834 AGAGGCAAAGAUUCACGCAUGGCUUCGGAUUGUGUCAAGUGCAAAGCAAACAAAACUUCAGCCAGCGACCCAAAAAGGGGCAAAUCAAUUU .((.((.........(((.(((((..(..((((......(((...))).))))..)...)))))))).(((.....)))))...))..... ( -22.50) >DroSec_CAF1 152288 91 + 1 AGAGGCAUAGAUUCACGCAUGGCUUCGGAGUAUGUCAAGUGCAAAGCAAACAAAACUUCAGCCAGCGACCCAAAAAGGGGCAAAUCAAUUU .((.((.........(((.(((((..(((((.(((....(((...))).)))..))))))))))))).(((.....)))))...))..... ( -24.90) >DroSim_CAF1 146220 91 + 1 AGGGGCAUAGAUUCACGCAUGGCUUCGGAUUGUGUCAAGUGCAAAGCAAACAAAACUUCAGCCAGCGACCCAAAAAGGGGCAAAUCAAUUU .((.((.........(((.(((((..(..((((......(((...))).))))..)...)))))))).(((.....)))))...))..... ( -21.60) >DroYak_CAF1 142940 90 + 1 AGAGGCAAAGAUUCACGCAUGGCUUCGGAUUGUGUCAAGUGCAAAACAAACAAAACUUCAGCCAGCAACCCAA-AAGGGGCAAAUCAAUUU .((.((..........((.(((((..(..((((.....((.....))..))))..)...)))))))..(((..-..)))))...))..... ( -19.30) >consensus AGAGGCAAAGAUUCACGCAUGGCUUCGGAUUGUGUCAAGUGCAAAGCAAACAAAACUUCAGCCAGCGACCCAAAAAGGGGCAAAUCAAUUU .((.((.........(((.(((((..((....(((....((.....)).)))...))..)))))))).(((.....)))))...))..... (-19.09 = -19.15 + 0.06)

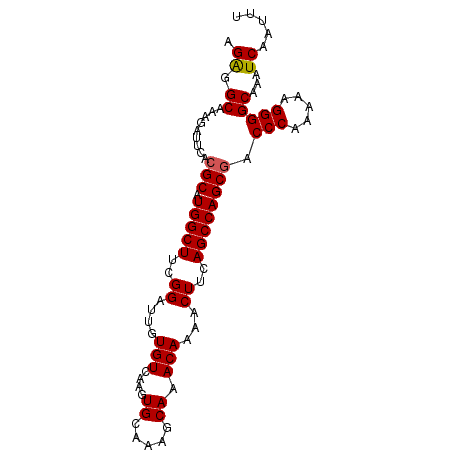

| Location | 17,301,676 – 17,301,767 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -22.29 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17301676 91 - 22407834 AAAUUGAUUUGCCCCUUUUUGGGUCGCUGGCUGAAGUUUUGUUUGCUUUGCACUUGACACAAUCCGAAGCCAUGCGUGAAUCUUUGCCUCU .....((((..((((.....)))..(((((((...(((.(((((((...)))...)))).)))....))))).)))..))))......... ( -23.30) >DroSec_CAF1 152288 91 - 1 AAAUUGAUUUGCCCCUUUUUGGGUCGCUGGCUGAAGUUUUGUUUGCUUUGCACUUGACAUACUCCGAAGCCAUGCGUGAAUCUAUGCCUCU .....((((..((((.....)))..(((((((..(((..(((((((...)))...)))).)))....))))).)))..))))......... ( -24.00) >DroSim_CAF1 146220 91 - 1 AAAUUGAUUUGCCCCUUUUUGGGUCGCUGGCUGAAGUUUUGUUUGCUUUGCACUUGACACAAUCCGAAGCCAUGCGUGAAUCUAUGCCCCU .....((((..((((.....)))..(((((((...(((.(((((((...)))...)))).)))....))))).)))..))))......... ( -23.30) >DroYak_CAF1 142940 90 - 1 AAAUUGAUUUGCCCCUU-UUGGGUUGCUGGCUGAAGUUUUGUUUGUUUUGCACUUGACACAAUCCGAAGCCAUGCGUGAAUCUUUGCCUCU .....((((..((((..-..)))..(((((((...(..((((..(((........)))))))..)..))))).)))..))))......... ( -24.20) >consensus AAAUUGAUUUGCCCCUUUUUGGGUCGCUGGCUGAAGUUUUGUUUGCUUUGCACUUGACACAAUCCGAAGCCAUGCGUGAAUCUAUGCCUCU .....((((..((((.....)))..(((((((...(((.(((((((...)))...)))).)))....))))).)))..))))......... (-22.29 = -22.35 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:19 2006