
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,295,750 – 17,295,925 |
| Length | 175 |
| Max. P | 0.610704 |

| Location | 17,295,750 – 17,295,861 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17295750 111 + 22407834 UUUUGCGGCUAAAUACGGUUAUGGCUUAUGGGAUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAACACCAACCCCGA-------UCCCCGCCGACGAGUCGUUUUUAA .((((((((........(((((......((((...........))))....)))))......(((((((................))-------))))))))).)))).......... ( -25.69) >DroSec_CAF1 146384 111 + 1 UUUUGCGGCUAAAUACGGUUAUGGCUUAUGGGCUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAACACCAUCCCCGA-------UCCCCGCCGACGCGUCGUUUUUAA ....(((((......(((..((((....(((((....)))))(((((.((........)).)))))...........))))..))).-------.....((....)))))))...... ( -29.60) >DroSim_CAF1 139823 111 + 1 UUUUGCGGCUAAAUACGGUUAUGGCUUAUGGGCUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAACACCAUCCCCGA-------UCCCCGCCGACGCGUCGUUUUUAA ....(((((......(((..((((....(((((....)))))(((((.((........)).)))))...........))))..))).-------.....((....)))))))...... ( -29.60) >DroEre_CAF1 133889 118 + 1 UUUUGCGGCUAAGUACGGUUAUGGCUUAUGGGCUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAACACCAUCCCCGAUCCCCAAUCCCCACCAACGCGCCGGUUUUAA .....((((...((..((((((((((....)))))))).))....................((((((((................))))))))..........))..))))....... ( -28.19) >DroYak_CAF1 137027 118 + 1 UUUUGCGGCUAAAUACGGUUAUGGCUUAUGGGCUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAGCACCAUCCCCGAUUCCCGAUCCCCGCCGACGCGUCGGUUUUAA .....(((...(((.(((..((((....(((((....)))))(((((.((........)).)))))...........))))..)))))).)))......(((((....)))))..... ( -32.30) >consensus UUUUGCGGCUAAAUACGGUUAUGGCUUAUGGGCUAUGGUCCACCCCAAAUAAUAACAAAUAUGGGGAUUUUACAACACCAUCCCCGA_______UCCCCGCCGACGCGUCGUUUUUAA .....((((......(((..((((....(((((....)))))(((((.((........)).)))))...........))))..))).............((....))))))....... (-24.26 = -24.90 + 0.64)

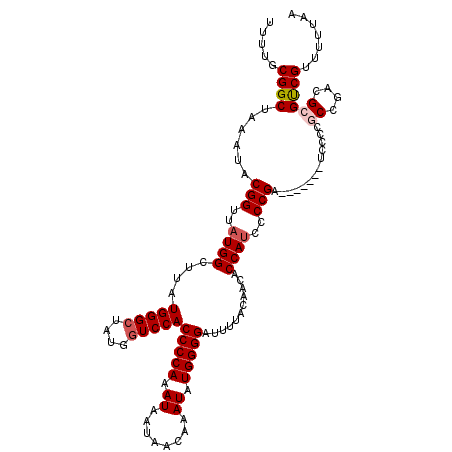

| Location | 17,295,828 – 17,295,925 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17295828 97 - 22407834 AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGAAGCAUUAAAAACGACUCGUCGGCGGGGA-------UCGGGGUUG ...................((((((((((((.(.(....((((((.......)))))).............(((....)))).).)))-------))))))))) ( -24.00) >DroSec_CAF1 146462 97 - 1 AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGGAGCAUUAAAAACGACGCGUCGGCGGGGA-------UCGGGGAUG ...((((((((((...)))).(((((.............((((((.......))))))................(((....)))))))-------).)))))). ( -25.50) >DroSim_CAF1 139901 97 - 1 AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGGAGCAUUAAAAACGACGCGUCGGCGGGGA-------UCGGGGAUG ...((((((((((...)))).(((((.............((((((.......))))))................(((....)))))))-------).)))))). ( -25.50) >DroEre_CAF1 133967 104 - 1 AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGGGGCAUUAAAACCGGCGCGUUGGUGGGGAUUGGGGAUCGGGGAUG ...((((((((((...))))..(((((((((.(((.(((((((((.......)))))..(.((..........)).)))))))).)))))))))...)))))). ( -34.50) >DroYak_CAF1 137105 104 - 1 AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGGGGCAUUAAAACCGACGCGUCGGCGGGGAUCGGGAAUCGGGGAUG ...((((((((((...))))..(((((((((.....(..((((((.......))))))..).........((((....))))...)))))))))...)))))). ( -36.50) >consensus AUAAUCCCCAGACGAAGUUUAAUCCCGAUUCACACACAAUGACAACGCACACUUGUCAGGAGCAUUAAAAACGACGCGUCGGCGGGGA_______UCGGGGAUG ...((((((((((...))))..(((((............((((((.......)))))).............(((....))).)))))..........)))))). (-23.66 = -23.38 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:14 2006