
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,294,185 – 17,294,288 |
| Length | 103 |
| Max. P | 0.715519 |

| Location | 17,294,185 – 17,294,288 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
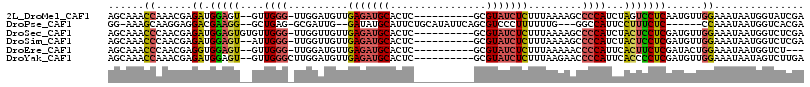
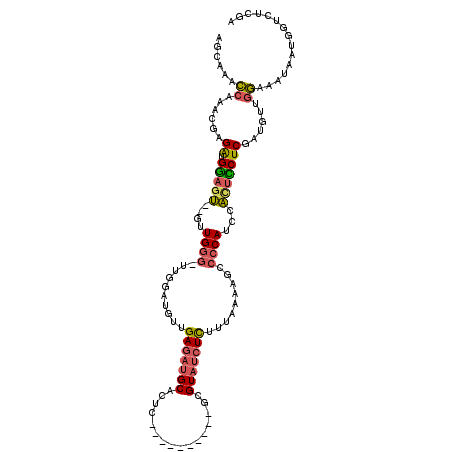
>2L_DroMel_CAF1 17294185 103 + 22407834 AGCAAACCAAACGAGAUGGAGU--GUUGGG-UUGGAUGUUGAGAUGCACUC----------GCGUAUCUCUUUAAAAGCCCCAUCUAGUCCUCAAUGUUGGAAAUAAUGGUAUCGA .....((((....((((((...--....((-((...((..(((((((....----------..)))))))..))..))))))))))..(((........))).....))))..... ( -26.01) >DroPse_CAF1 212935 101 + 1 GG-AAAGCAAGGAGGACGAAGG--GCUGAG-GCGAUUG--GAUAUGCAUUCUGCAUAUUCAGCGUCCCUUUUUUG---GGCCAUUCCUUUCUC------CCAAAUAAUGGUCACGA ..-.......((((((.(((.(--((...(-(((.(((--(((((((.....))))))))))))))((......)---)))).)))..)))))------)................ ( -32.40) >DroSec_CAF1 144808 105 + 1 AGCAAACCCAACGAGAUGGAGUGUGUUGGG-UUGGUUGUUGAGAUGCACUC----------GCGUAUCUCUUUAAAAGCCCCAUCUACUCCUCGAUGUUGGAAAUAAUGGUCUCGA .......((((((.((.((((((.((.(((-((..(((..(((((((....----------..)))))))..))).))))).)).))))))))..))))))............... ( -37.00) >DroSim_CAF1 138249 103 + 1 AGCAAACCCAACGAGAUGGAGU--AUUGGG-UUGGUUGUUGAGAUGCACUC----------GCGUAUCUCUUUAAAAGCCCCAUCUACUCCUCGAUGUUGGAAAUAAUGGUCUCGA .......((((((.((.(((((--(..(((-((..(((..(((((((....----------..)))))))..))).)))))....))))))))..))))))............... ( -35.00) >DroEre_CAF1 132575 100 + 1 AGCAAACCCAACGAGGUGGAGU--GUUGGG-UUGGAUGUUGAGAUGCACUC----------GCGUAUCUCUUUAAAAACCCCAUUCACUUCUCGAUACUGGAAAUAAUGGUCU--- .......(((.(((((..((((--(..(((-((...((..(((((((....----------..)))))))..))..))))))))))...)))))....)))............--- ( -29.30) >DroYak_CAF1 135506 104 + 1 AGCAAACCAAACGAGAUGGAGU--GUUGGGCUUGGAUGUUGAGAUGCACUC----------GCGUAUCUCUUUAAGAACCCCAUUCACCCCUCGAUGUUGGAAAUAAUAGUCUUGA ......((((.((((..((.((--(.((((((((((....(((((((....----------..)))))))))))))...))))..)))))))))...))))............... ( -29.60) >consensus AGCAAACCAAACGAGAUGGAGU__GUUGGG_UUGGAUGUUGAGAUGCACUC__________GCGUAUCUCUUUAAAAGCCCCAUCCACUCCUCGAUGUUGGAAAUAAUGGUCUCGA ......((......((.(((((....((((..........(((((((................))))))).........))))...)))))))......))............... (-16.30 = -16.33 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:11 2006