
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,288,983 – 17,289,231 |
| Length | 248 |
| Max. P | 0.980991 |

| Location | 17,288,983 – 17,289,095 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -31.70 |
| Energy contribution | -31.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17288983 112 + 22407834 GACGAAAACGUGGAAAAUGGGGGAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUCGCUCUGUGGCAGCUGCCA .(((....)))...................((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). ( -32.40) >DroSec_CAF1 139072 112 + 1 GACGAAAACGAGGAAAAUGGGGGAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCA ..............................((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). ( -31.70) >DroSim_CAF1 132228 112 + 1 GACGAAAACGUGGAAAAUGGGGGAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCA .(((....)))...................((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). ( -32.40) >DroEre_CAF1 127323 111 + 1 GACGA-AACGUGGGAAAUGGGGGAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCA .....-..(((.....)))...........((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). ( -32.10) >DroYak_CAF1 130096 111 + 1 GAAGAAAACGUGGGAAAUGG-GAAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCA ........(((.....))).-.........((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). ( -32.10) >consensus GACGAAAACGUGGAAAAUGGGGGAAGUGGGGGCGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCA ..............................((((((.......((.(..((.....((((.((....)).)))).))..).)).((..(((......)))..)).)))))). (-31.70 = -31.70 + -0.00)

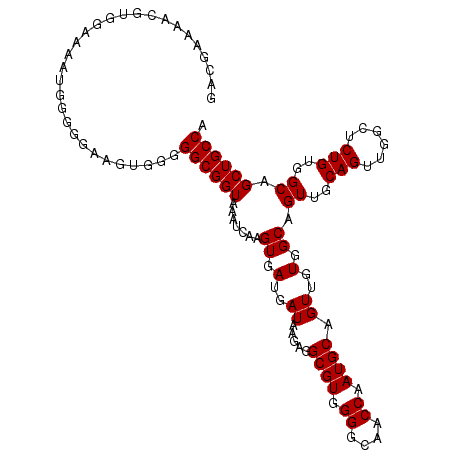
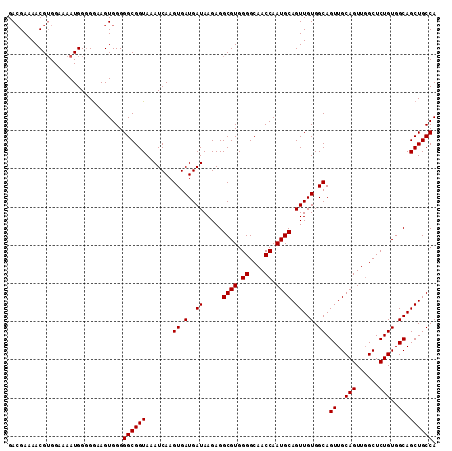
| Location | 17,289,015 – 17,289,134 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -38.52 |
| Energy contribution | -38.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17289015 119 + 22407834 CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUCGCUCUGUGGCAGCUGCCACUUCAAAUAACUUCACUGCACACCGUUUUUCCCGCUUUC ((((......(((((.(.......((((.((....)).)))).(..(((((((((((.............)))))))))))..)......).)))))....)))).............. ( -38.52) >DroSec_CAF1 139104 119 + 1 CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUC ((((......(((((.(.......((((.((....)).)))).(..(((((((((((.............)))))))))))..)......).)))))....)))).............. ( -38.52) >DroSim_CAF1 132260 119 + 1 CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUC ((((......(((((.(.......((((.((....)).)))).(..(((((((((((.............)))))))))))..)......).)))))....)))).............. ( -38.52) >DroEre_CAF1 127354 119 + 1 CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUGCCGCUUUC (((((((....(((.((...((.((.(((((....))...((((((((.((((..((.....)).)))).))))))))..........))).)).)).)))))....)))))))..... ( -41.80) >DroYak_CAF1 130127 119 + 1 CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUC ((((......(((((.(.......((((.((....)).)))).(..(((((((((((.............)))))))))))..)......).)))))....)))).............. ( -38.52) >consensus CGGUAAAUCAAGUGAUGAUAAGAGGCGUGGGGCAACCAAUGCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUC ((((......(((((.(.......((((.((....)).)))).(..(((((((((((.............)))))))))))..)......).)))))....)))).............. (-38.52 = -38.52 + -0.00)

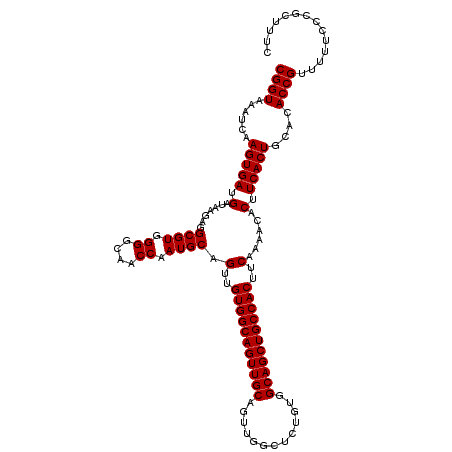

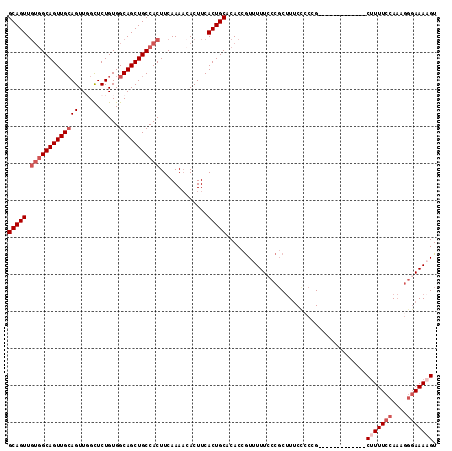
| Location | 17,289,055 – 17,289,158 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -25.31 |
| Energy contribution | -26.64 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17289055 103 + 22407834 GCAGUUGUGGCAGUUGCAGUUCGCUCUGUGGCAGCUGCCACUUCAAAUAACUUCACUGCACACCGUUUUUCCCGCUUUCUCCCG-------------CUUUUCCAAAGGGAAAAGU (((((.(((((((((((.............))))))))))).............)))))........................(-------------(((((((....)))))))) ( -32.66) >DroSec_CAF1 139144 103 + 1 GCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCACCG-------------CUUUUCCAAAGGGAAAAGU (((((.(((((((((((.............))))))))))).............)))))........................(-------------(((((((....)))))))) ( -32.66) >DroSim_CAF1 132300 103 + 1 GCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCCG-------------CUUUUCCAAAGGGAAAAGU (((((.(((((((((((.............))))))))))).............)))))........................(-------------(((((((....)))))))) ( -32.66) >DroEre_CAF1 127394 103 + 1 GCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUGCCGCUUUCCCCGG-------------CUUUUCCAAAGGGAAAUGU (((((.(((((((((((.............))))))))))).............)))))...........((((.......)))-------------).(((((....)))))... ( -32.76) >DroYak_CAF1 130167 103 + 1 GCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCGG-------------CUUUUCCAAAGGGAAAAGU (((((.(((((((((((.............))))))))))).............)))))........................(-------------(((((((....)))))))) ( -32.76) >DroPer_CAF1 199845 109 + 1 GCAGUU---GCAGUUGGAGUUCGUUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACAAUUUUUUCACCCUCCCCCUGCCCCCCCGCCUGCCAUUUCCAC----AAACGU ((((.(---(((((.(((((.......(((((....)))))........))))))))))).......................((......)))))).........----...... ( -25.66) >consensus GCAGUUGUGGCAGUUGCAGUUGGCUCUGUGGCAGCUGCCACUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCCG_____________CUUUUCCAAAGGGAAAAGU (((((.(((((((((((((......))...))))))))))).............)))))......................................(((((((....))))))). (-25.31 = -26.64 + 1.33)


| Location | 17,289,095 – 17,289,192 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.38 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17289095 97 + 22407834 CUUCAAAUAACUUCACUGCACACCGUUUUUCCCGCUUUCUCCCGCUUUUCCAAAGGGAAAAGUGAGGGAGUGCCGCAUGUGAAGAGGGGCUAGAGGU ((((......(((((((((.....(.....).((((..(((..((((((((....)))))))))))..))))..))).))))))........)))). ( -29.94) >DroSec_CAF1 139184 97 + 1 CUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCACCGCUUUUCCAAAGGGAAAAGUGAGGGAGUGCCGCAUGUGAAGAGGGGCUAGAGGU ((((.(..(.(((((((((.....(.......)((.((((..(((((((((....)))))))))..)))).)).))).))))))...)..).)))). ( -31.90) >DroSim_CAF1 132340 97 + 1 CUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCCGCUUUUCCAAAGGGAAAAGUGAGGGAGUGCCGCAUGUGAAGAGGGGCUAGAGGU ((((.(..(.(((((((((.....(.......)((.(((((.(((((((((....))))))))).))))).)).))).))))))...)..).)))). ( -33.90) >DroEre_CAF1 127434 97 + 1 CUUCAAAACACUUCACUGCACACCGUUUUUGCCGCUUUCCCCGGCUUUUCCAAAGGGAAAUGUGGGGGAGUGCCGCAUGUGAAGAGGGGCUAGAGGU .........(((((...((...((.(((.(((.((.((((((.((.(((((....))))).)).)))))).)).)))...)))..)).))..))))) ( -35.60) >DroYak_CAF1 130207 97 + 1 CUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCGGCUUUUCCAAAGGGAAAAGUGAGGGAUUGCCGCAUAUGAAGAGGGGCUAGAGGU .........(((((...((...((.((((..(.((..((((..((((((((....))))))))..))))..)).).....)))).)).))..))))) ( -29.10) >consensus CUUCAAAACACUUCACUGCACACCGUUUUUCCCGCUUUCCCCCGCUUUUCCAAAGGGAAAAGUGAGGGAGUGCCGCAUGUGAAGAGGGGCUAGAGGU ((((....(.(((((((((.....(.......)((.(((((.(((((((((....))))))))).))))).)).))).)))))).)......)))). (-28.14 = -29.38 + 1.24)

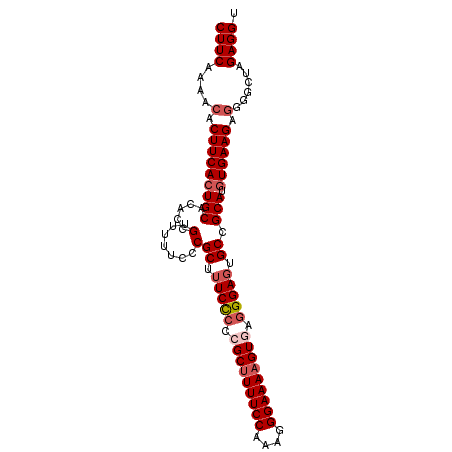

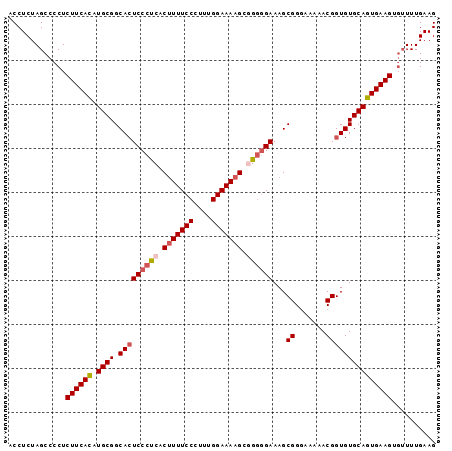
| Location | 17,289,095 – 17,289,192 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.54 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17289095 97 - 22407834 ACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGAGAAAGCGGGAAAAACGGUGUGCAGUGAAGUUAUUUGAAG .............((((((.((((.(((((((.(..((((((((((....))).))))))).).))))......))))))))))))).......... ( -28.60) >DroSec_CAF1 139184 97 - 1 ACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGUGGAAAGCGGGAAAAACGGUGUGCAGUGAAGUGUUUUGAAG ...((.(((....((((((.((((.((((((.((.(((((((....))))))).)).)))...((.......))))))))))))))).)))..)).. ( -28.80) >DroSim_CAF1 132340 97 - 1 ACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGGGAAAGCGGGAAAAACGGUGUGCAGUGAAGUGUUUUGAAG ...((.(((....((((((.((((.(((((((((.(((((((....))))))).))))))...((.......))))))))))))))).)))..)).. ( -35.30) >DroEre_CAF1 127434 97 - 1 ACCUCUAGCCCCUCUUCACAUGCGGCACUCCCCCACAUUUCCCUUUGGAAAAGCCGGGGAAAGCGGCAAAAACGGUGUGCAGUGAAGUGUUUUGAAG ...((.(((....((((((.((((.((((((((..(.(((((....))))).)..)))))...((.......))))))))))))))).)))..)).. ( -29.20) >DroYak_CAF1 130207 97 - 1 ACCUCUAGCCCCUCUUCAUAUGCGGCAAUCCCUCACUUUUCCCUUUGGAAAAGCCGGGGAAAGCGGGAAAAACGGUGUGCAGUGAAGUGUUUUGAAG ...((.(((....((((((.((((....(((((..(((((((....)))))))..)))))..((.(......).)).)))))))))).)))..)).. ( -26.70) >consensus ACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGGGAAAGCGGGAAAAACGGUGUGCAGUGAAGUGUUUUGAAG .............((((((.((((.(((((((((.(((((((....))))))).))))))...((.......))))))))))))))).......... (-28.66 = -29.54 + 0.88)


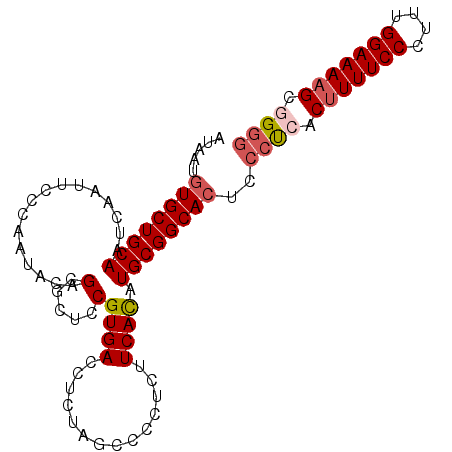
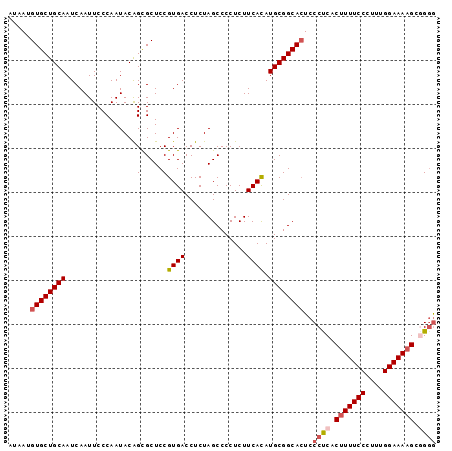
| Location | 17,289,134 – 17,289,231 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17289134 97 - 22407834 AUAAUGUGCUGCAAUCAAUUCCCAAUACAGCGCUCCGUGACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGA .....((((((((................(.....)((((..............)))).))))))))((((...(((((((....))))))).)))) ( -24.54) >DroSec_CAF1 139223 97 - 1 AUAAUGUGCUGCAAUCAAUUCCCAAUACAGCGCUCCGUGACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGUG .....((((((((................(.....)((((..............)))).))))))))....((.(((((((....))))))).)).. ( -20.74) >DroSim_CAF1 132379 97 - 1 AUAAUGUGCUGCAAUCAAUUCCCAAUACAGCGCUCCGUGACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGG .....((((((((................(.....)((((..............)))).))))))))..((((.(((((((....))))))).)))) ( -26.24) >DroEre_CAF1 127473 97 - 1 AUAAUGUGCUGCAAUCAAUUCCCAAUACGGCGCUCCGUGACCUCUAGCCCCUCUUCACAUGCGGCACUCCCCCACAUUUCCCUUUGGAAAAGCCGGG .....((((((((...............((....))((((..............)))).))))))))..(((..(.(((((....))))).)..))) ( -23.34) >DroYak_CAF1 130246 97 - 1 AUAAUGUGCUGCAAUCAAUUCCCAAUACGGUGCUCCGUGACCUCUAGCCCCUCUUCAUAUGCGGCAAUCCCUCACUUUUCCCUUUGGAAAAGCCGGG ......(((((((............(((((....)))))..(....)............)))))))..(((...(((((((....)))))))..))) ( -23.30) >consensus AUAAUGUGCUGCAAUCAAUUCCCAAUACAGCGCUCCGUGACCUCUAGCCCCUCUUCACAUGCGGCACUCCCUCACUUUUCCCUUUGGAAAAGCGGGG .....((((((((................(.....)((((..............)))).))))))))..((((.(((((((....))))))).)))) (-20.64 = -21.52 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:03 2006