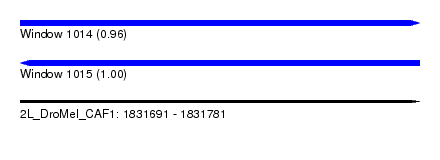
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,831,691 – 1,831,781 |
| Length | 90 |
| Max. P | 0.998724 |

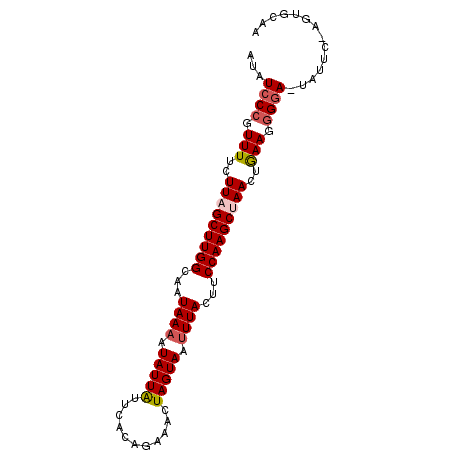
| Location | 1,831,691 – 1,831,781 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1831691 90 + 22407834 UUGCACU-GAAUA-UCCCCUUCAGUUAGCUUGGAAGUAAAUUACUAGUUUCUGUGAAUAAUAUUUUAUUGCCAAGCUAAGAAAACGGGAUAU .......-..(((-((((.(((..(((((((((.((((((((((........)))).......)))))).))))))))))))...))))))) ( -24.31) >DroSec_CAF1 51695 90 + 1 UUGCACU-GAAUA-UCCCCUUCAGUUAGCUUGGAAGUAAGUUACUAGUUUCUGUGAAUAAUAUUUUAUUGCCAAGCAAAGAAAACGGGAUAU .......-..(((-((((.........((((((.((((((((((........)))).......)))))).)))))).........))))))) ( -19.98) >DroSim_CAF1 32261 91 + 1 UUGCACU-GAAUAAUUCCCUUCAGUUAGCUUGGAAGUAAAUUACUAGUUUCUGUGAAUAAUAUUUUAUUGCCAAGCUAAGAAAACGGGAUAU .......-.......(((((((..(((((((((.((((((((((........)))).......)))))).))))))))))))...))))... ( -21.01) >DroEre_CAF1 42135 87 + 1 --GCAGU-GAAUA-UCCCCUUUAGUUGGCUUGGAAUUAAAUUACUAGU-UCGAUGAAUAAUAUUUUAUUGCCAAGCUAAGAAAAUGGGAUAU --.....-..(((-((((.(((..((((((((((((((......))))-)(((((((......))))))))))))))))..))).))))))) ( -25.90) >DroYak_CAF1 16710 89 + 1 --GUUGUAGAAUA-UCCACUUCAGUUAGCUUGGAAGUAAAUUACUAGUUUCAAUCAACAAUAUUUUAUUGCCAAGCUAACAAAAUGGGAUAU --........(((-(((......((((((((((.((((((.((...(((......)))..)).)))))).))))))))))......)))))) ( -24.60) >consensus UUGCACU_GAAUA_UCCCCUUCAGUUAGCUUGGAAGUAAAUUACUAGUUUCUGUGAAUAAUAUUUUAUUGCCAAGCUAAGAAAACGGGAUAU ..............((((......(((((((((.((((((.......................)))))).)))))))))......))))... (-16.78 = -16.94 + 0.16)

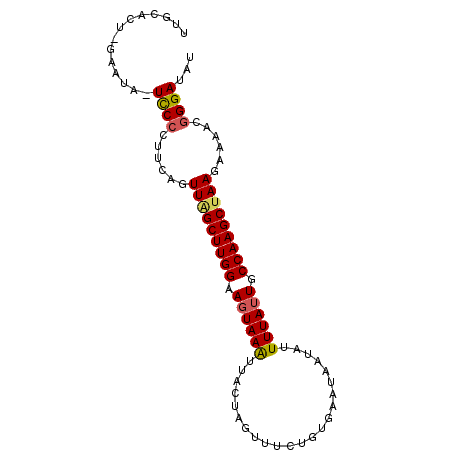
| Location | 1,831,691 – 1,831,781 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1831691 90 - 22407834 AUAUCCCGUUUUCUUAGCUUGGCAAUAAAAUAUUAUUCACAGAAACUAGUAAUUUACUUCCAAGCUAACUGAAGGGGA-UAUUC-AGUGCAA (((((((.(((..(((((((((...((((.((((((((...)))..))))).))))...)))))))))..))).))))-)))..-....... ( -26.90) >DroSec_CAF1 51695 90 - 1 AUAUCCCGUUUUCUUUGCUUGGCAAUAAAAUAUUAUUCACAGAAACUAGUAACUUACUUCCAAGCUAACUGAAGGGGA-UAUUC-AGUGCAA (((((((.(((..((.((((((...(((..((((((((...)))..)))))..)))...)))))).))..))).))))-)))..-....... ( -20.60) >DroSim_CAF1 32261 91 - 1 AUAUCCCGUUUUCUUAGCUUGGCAAUAAAAUAUUAUUCACAGAAACUAGUAAUUUACUUCCAAGCUAACUGAAGGGAAUUAUUC-AGUGCAA ...((((.(((..(((((((((...((((.((((((((...)))..))))).))))...)))))))))..))))))).......-....... ( -21.70) >DroEre_CAF1 42135 87 - 1 AUAUCCCAUUUUCUUAGCUUGGCAAUAAAAUAUUAUUCAUCGA-ACUAGUAAUUUAAUUCCAAGCCAACUAAAGGGGA-UAUUC-ACUGC-- (((((((.(((..((.((((((...((((.((((((((...))-).))))).))))...)))))).))..))).))))-)))..-.....-- ( -21.00) >DroYak_CAF1 16710 89 - 1 AUAUCCCAUUUUGUUAGCUUGGCAAUAAAAUAUUGUUGAUUGAAACUAGUAAUUUACUUCCAAGCUAACUGAAGUGGA-UAUUCUACAAC-- ((((((.(((((((((((((((...((((.(((((((......))).)))).))))...)))))))))).))))))))-)))........-- ( -26.60) >consensus AUAUCCCGUUUUCUUAGCUUGGCAAUAAAAUAUUAUUCACAGAAACUAGUAAUUUACUUCCAAGCUAACUGAAGGGGA_UAUUC_AGUGCAA ...((((.(((..(((((((((...((((.(((((...........))))).))))...)))))))))..))).)))).............. (-18.42 = -19.10 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:10 2006