| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,287,414 – 17,287,532 |
| Length | 118 |
| Max. P | 0.832676 |

| Location | 17,287,414 – 17,287,532 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
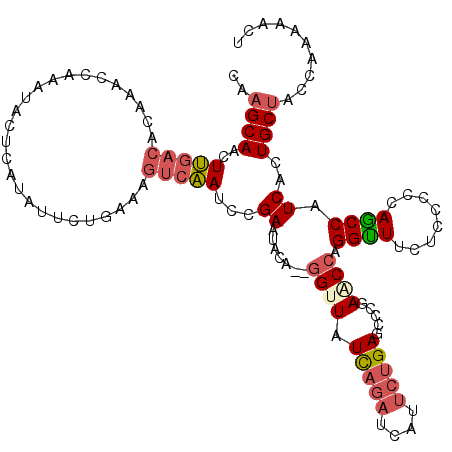
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -12.31 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
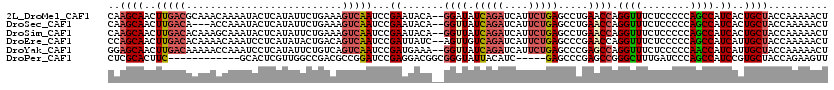
>2L_DroMel_CAF1 17287414 118 - 22407834 CAAGCAACUUGACGCAAACAAAAUACUCAUAUUCUGAAAGUCAAUCCGAAUACA--GGAUAUCAGAUCAUUCUGAGCCUGAACCAGGUUUCUCCCCCAGCCAUCACUGCUACCAAAAACU ..((((..(((((.....((.((((....)))).))...)))))...((...((--((...(((((....))))).)))).....((((........)))).))..)))).......... ( -23.30) >DroSec_CAF1 137529 115 - 1 CAAGCAACUUGACA---ACCAAAUACUCAUAUUCUGAAAGUCAAUCCGAAUACA--GGUUAUCAGAUCAUUCUGAGCCUGAACCAGGUUUCUCCCCCAGCCAUCACUGCUACCAAAAACU ..((((..(((((.---..((((((....)))).))...)))))...((...((--(((..(((((....)))))))))).....((((........)))).))..)))).......... ( -22.20) >DroSim_CAF1 130673 118 - 1 CAAGCAACUUGACACAAAGCAAAUACUCAUAUUCUGAAAGUCAAUCCGAAUACA--GGUUAUCAGAUCAUUCUGAGCCUGAACCAGGUUUCUCCCCCAGCCAUCACUGCUACCAAAAACU ..((((..(((((....((..((((....))))))....)))))...((...((--(((..(((((....)))))))))).....((((........)))).))..)))).......... ( -21.90) >DroEre_CAF1 125773 118 - 1 CCAGCAACUUGACACAAAACAAAUCCUCAUAUACUGACAGUCAAUCCGAUUAUC--AGUUGUCAGAUCAUUCUGAGCCCGAACCAGGUUUCUCCCCCAGCCAUCAUUGCUACCAAAAACU ..(((((..(((..............((....(((((.((((.....)))).))--)))..(((((....)))))....))....((((........)))).)))))))).......... ( -20.60) >DroYak_CAF1 128575 118 - 1 GGAGCAACUUGACAAAAACCAAAUCCUCAUAUUCUGUCAGUCAAUCCGAUGAAA--GGUUAUCAGAUCAUUCUGAGCCCGAGCCAGGUUUCUCCCCCAACCAUCAUUGCUACCAAAAACU (((((((.((((((....................)))))).......((((...--((((.(((((....))))).....)))).((........))...)))).))))).))....... ( -25.25) >DroPer_CAF1 198575 103 - 1 CUCGCACUUC------------GCACUCGUUGGCCGACGCCGGAUCCGAGGACGGCGGGUAUUACAUC-----GAGCCCGAGCCGGGCUUUGAUCCCAGCCAUCCGUGCUACCAGAAGUU .....(((((------------((((((((((((....)))))...)))(((.((((((.((((....-----(((((((...)))))))))))))).))).))))))).....))))). ( -39.40) >consensus CAAGCAACUUGACACAAACCAAAUACUCAUAUUCUGAAAGUCAAUCCGAAUACA__GGUUAUCAGAUCAUUCUGAGCCCGAACCAGGUUUCUCCCCCAGCCAUCACUGCUACCAAAAACU ..((((..(((((..........................)))))...((.......((((.(((((....))))).....)))).((((........)))).))..)))).......... (-12.31 = -13.20 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:56 2006